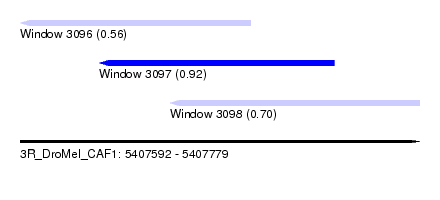
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,407,592 – 5,407,779 |
| Length | 187 |
| Max. P | 0.915062 |

| Location | 5,407,592 – 5,407,700 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -39.59 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5407592 108 - 27905053 ACGUUUCCAUGACGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCA------AUGGCG-AUGACAACGAUGGCG---AUAUGUGGCCAAUGCAACUGCAACUGCAA ............((((-((((((.......))))-))))))....(((.(((((((((..------.((((.-((.....((....))---....)).)))))))).)))))....))). ( -38.80) >DroSec_CAF1 11528 111 - 1 ACGUUUCCAUGGCGUUAGGCCGCAAACAAAGUGGUCCAGCGAAAAUGCGGCAGUGCAUCA------ACGGCGAAUGACAACGUUGGCA---AUAUGCGGCCAUGCCAACUGCAACUGCAA ..(((..(((((((((.((((((.......)))))).))).........(((.(((..((------(((...........))))))))---...))).))))))..)))(((....))). ( -36.10) >DroSim_CAF1 6480 108 - 1 ACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCA------ACGGCG-AUGACAACGUUGGCG---AUAUGCGGCCAAUGCAACUGCAACUGCAA .(((........((((-((((((.......))))-)))))).....)))((((((((...------.((.((-(((....))))).))---...((((.....))))..))).))))).. ( -41.42) >DroEre_CAF1 10775 114 - 1 ACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCAAAAUCAAAAGCG-GUGACAACGAUGGCA---AUAUGUGGCUAAUGCAACUGCAACUGCAA .(((........((((-((((((.......))))-)))))).....)))(((((((((.............(-....).....((((.---.......)))))))).)))))........ ( -37.02) >DroYak_CAF1 6591 111 - 1 ACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCA------AAAGCG-GCGACAACGUUGGCAACAAUAUGUGGCCAAUGCAACUGCAACUGCAA .(((........((((-((((((.......))))-)))))).....)))(((((((....------...))(-((.(((..((((....)))).))).)))..(((....)))))))).. ( -44.62) >consensus ACGUUUCCAUGGCGUU_GGCCGCAAACAAAGUGG_CCAGCGAAAAUGCGGCAGUGCAUCA______AAGGCG_AUGACAACGUUGGCA___AUAUGUGGCCAAUGCAACUGCAACUGCAA .(((........((((.((((((.......)))).)))))).....)))((((((((...........((((........))))(((...........)))........))).))))).. (-25.80 = -25.80 + 0.00)


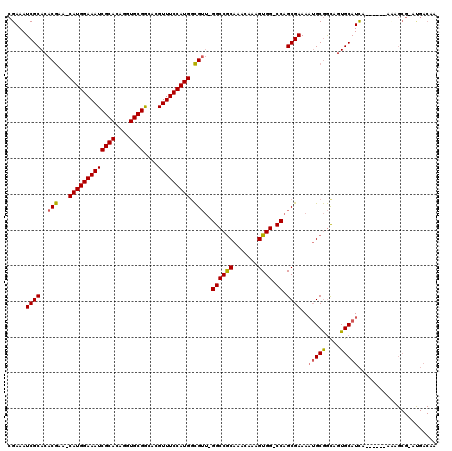
| Location | 5,407,629 – 5,407,739 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -31.79 |
| Energy contribution | -31.73 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5407629 110 - 27905053 CGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGACGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCA------AUGGCG-AUGACAA .....(((((......-(((((((((((((....))))....))))))))).((((-((((((.......))))-))))))....)))))..((.((((.------.....)-))))).. ( -44.00) >DroSec_CAF1 11565 114 - 1 CGAAAUCGCACACGAACCAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUUAGGCCGCAAACAAAGUGGUCCAGCGAAAAUGCGGCAGUGCAUCA------ACGGCGAAUGACAA .....((((.......((((((((((((((....))))....))))))))))((((.((((((.......)))))).))))...(((((....)))))..------...))))....... ( -38.70) >DroSim_CAF1 6517 110 - 1 CGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCA------ACGGCG-AUGACAA .....(((((......-(((((((((((((....))))....))))))))).((((-((((((.......))))-))))))....)))))..((.((((.------.....)-))))).. ( -43.90) >DroEre_CAF1 10812 116 - 1 CGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCAAAAUCAAAAGCG-GUGACAA .(..(((((.......-(((((((((((((....))))....))))))))).((((-((((((.......))))-))))))...(((((....)))))...........)))-))..).. ( -43.20) >DroYak_CAF1 6631 110 - 1 CGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGCGAAAAUGCGGCAGUGCAUCA------AAAGCG-GCGACAA .....((((.......-(((((((((((((....))))....)))))))))(((((-((((((.......))))-)))))....(((((....)))))..------...)).-))))... ( -43.00) >DroAna_CAF1 6352 99 - 1 CGAAAUCGCAGACAAA-CAUGGAAAUCGCACAGGUGCGACACGUUUCCAUGGUGGC-GGCCACAAGU-CAGUGG-CCAGCGAAACGGCAAUAAUGCAUUA------AUA----------- ((...((((.......-(((((((((((((....)))))....)))))))).....-((((((....-..))))-)).))))..))(((....)))....------...----------- ( -36.70) >consensus CGAAAUCGCACACGAA_CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU_GGCCGCAAACAAAGUGG_CCAGCGAAAAUGCGGCAGUGCAUCA______AAAGCG_AUGACAA .....((((..(((...(((((((((((((....))))....))))))))).)))..((((((.......)))).)).))))..(((((....)))))...................... (-31.79 = -31.73 + -0.05)
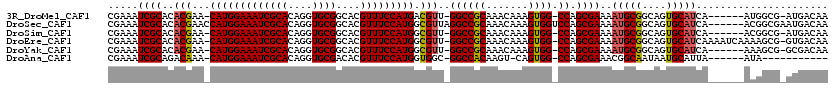
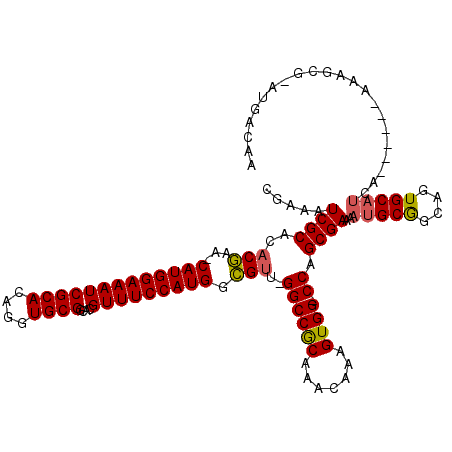
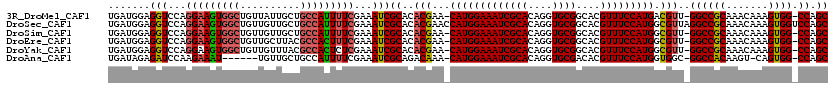
| Location | 5,407,662 – 5,407,779 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -35.73 |
| Energy contribution | -35.82 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
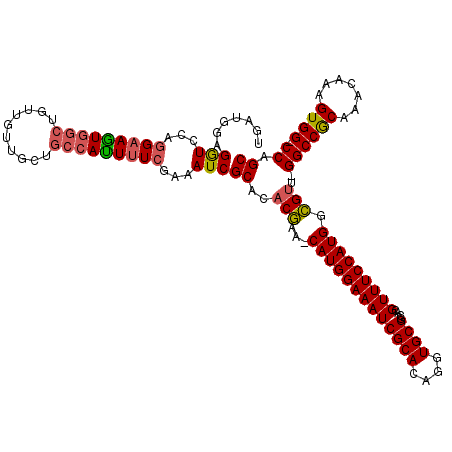
>3R_DroMel_CAF1 5407662 117 - 27905053 UGAUGGAGGUCCAGGAAGUGGCUGUUAUUGCUGCCAUUUUCGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGACGUU-GGCCGCAAACAAAGUGG-CCAGC ...(((....)))(((((((((.((....)).)))))))))....(((....))).-(((((((((((((....))))....)))))))))..(((-((((((.......))))-))))) ( -47.60) >DroSec_CAF1 11599 120 - 1 UGAUGGAGGUCCAGGAAGUGGCUGUUGUUGCUGCCAUUUUCGAAAUCGCACACGAACCAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUUAGGCCGCAAACAAAGUGGUCCAGC ...(((....)))(((((((((.((....)).)))))))))....(((....))).((((((((((((((....))))....)))))))))).(((.((((((.......)))))).))) ( -44.30) >DroSim_CAF1 6550 117 - 1 UGAUGGAGGUCCAGGAAGUGGCUGUUGUUGCUGCCAUUUUCGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGC ...(((....)))(((((((((.((....)).)))))))))....(((....))).-(((((((((((((....))))....)))))))))..(((-((((((.......))))-))))) ( -47.80) >DroEre_CAF1 10851 117 - 1 UGAUGGAGGUCCAGGAAGUGGCUGUUGCUUACGCCACUUUCGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGC ...(((....)))(((((((((.((.....)))))))))))....(((....))).-(((((((((((((....))))....)))))))))..(((-((((((.......))))-))))) ( -48.80) >DroYak_CAF1 6664 117 - 1 UGAUGGAGGUCCAGGAAGUGGCUGUUGUUUACGCCACUCUCGAAAUCGCACACGAA-CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU-GGCCGCAAACAAAGUGG-CCAGC ...(((....)))(..((((((.((.....))))))))..)....(((....))).-(((((((((((((....))))....)))))))))..(((-((((((.......))))-))))) ( -46.20) >DroAna_CAF1 6375 110 - 1 UGAUAGAGAUCCAAGAAAU------UGUUGCUGCCAUUUUCGAAAUCGCAGACAAA-CAUGGAAAUCGCACAGGUGCGACACGUUUCCAUGGUGGC-GGCCACAAGU-CAGUGG-CCAGC ...........(((....)------))..(((((((((((((....)).)))....-(((((((((((((....)))))....)))))))))))))-((((((....-..))))-))))) ( -38.30) >consensus UGAUGGAGGUCCAGGAAGUGGCUGUUGUUGCUGCCAUUUUCGAAAUCGCACACGAA_CAUGGAAAUCGCACAGGUGCGGCACGUUUCCAUGGCGUU_GGCCGCAAACAAAGUGG_CCAGC .......(((...(((((((((..........)))))))))...)))((..(((...(((((((((((((....))))....))))))))).)))..((((((.......)))).)).)) (-35.73 = -35.82 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:56 2006