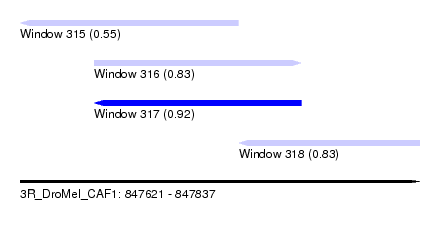
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 847,621 – 847,837 |
| Length | 216 |
| Max. P | 0.924223 |

| Location | 847,621 – 847,739 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -22.24 |
| Energy contribution | -21.43 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 847621 118 - 27905053 --AUACAUACGUACUACAACCCAGACUCGCCGAUUCUUCGCCCUCGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCAUAACGAGGUCCGAACUAUCCGCACUGUGCUCCAGUCCGCC --........((((....((((((((..(.(((....))).)...))))))))......((...(((.((((..((((.....))))..)))))))....))...))))........... ( -31.20) >DroSec_CAF1 23291 120 - 1 ACAUACAUACAUACUGCAGCCCAGACUCGCCGAUUCUUCACCCUCGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCAUAACGAGGUCCAAACUAUCCGCACUAUGCCACAGCCCACC .........((((.(((.(((.((((((((.((.....(((((......)))))..)).))..)))))).))).((((.....)))).............))).))))............ ( -31.60) >DroSim_CAF1 22539 120 - 1 ACAUACAUACAUACUACAGCCCAGACUCGCCGAUUCUUCACCCUUGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCAUAACGAGGUCCAAACUAUCCGCACUAUGCCACAGCCCACC ..................(((.((((((((.((.....(((((......)))))..)).))..)))))).)))....(((((....((..........))....)))))........... ( -28.90) >DroEre_CAF1 24351 108 - 1 ACGUAC--UCGUACUACCACCCAGACUCGCCGGCUCCUCGUCUUUGUCUGGGUGUUUCAGCCGGAGUCUCGGUAUGCGCAUAACGAGCUUCAAACUAUCC----------ACAGCCCACC ..(((.--...)))...(((((((((.....(((.....)))...)))))))))........((((.((((.(((....))).)))))))).........----------.......... ( -29.00) >consensus ACAUACAUACAUACUACAACCCAGACUCGCCGAUUCUUCACCCUCGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCAUAACGAGGUCCAAACUAUCCGCACUAUGCCACAGCCCACC ..................(((.((((((((.((.....(((((......)))))..)).))..)))))).))).((((.....))))................................. (-22.24 = -21.43 + -0.81)

| Location | 847,661 – 847,773 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -30.51 |
| Energy contribution | -31.45 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 847661 112 + 27905053 UGCGAAUACCGAGACUCUGGCUGAAACACCCAGACGAGGGCGAAGAAUCGGCGAGUCUGGGUUGUAGUACGUAUGUAU--------GGUGGAUUCGGCACAAGUGUGCUAUGUACAGCUC .((....(((.((((((..(((((..(.(((......))).).....))))))))))).)))....(((((((.((((--------.(((.......)))....))))))))))).)).. ( -36.40) >DroSec_CAF1 23331 120 + 1 UGCGAAUACCGAGACUCUGGCUGAAACACCCAGACGAGGGUGAAGAAUCGGCGAGUCUGGGCUGCAGUAUGUAUGUAUGUAUGUGAGGUGGAUUCGGCACAAGUGUGCUGUGUACAGCUC .((((((.((.((((((..(((((..(((((......))))).....))))))))))).))(..(..(((((((....)))))))..)..)))))(((((....))))).......)).. ( -43.50) >DroSim_CAF1 22579 120 + 1 UGCGAAUACCGAGACUCUGGCUGAAACACCCAGACAAGGGUGAAGAAUCGGCGAGUCUGGGCUGUAGUAUGUAUGUAUGUAUGUGAGGUGGAUUCGGCACAAGUGUGCUAUGUACAGCUC .((..(((((.((((((..(((((..(((((......))))).....))))))))))).)).))).(((((((.(((..(.((((.((.....))..)))).)..)))))))))).)).. ( -42.20) >DroEre_CAF1 24381 118 + 1 UGCGCAUACCGAGACUCCGGCUGAAACACCCAGACAAAGACGAGGAGCCGGCGAGUCUGGGUGGUAGUACGA--GUACGUAUGUACGCUGGAUUCGGCACAAGUGUGCUAUGUGCAGCUC ((((((((.((((..((((((.....(((((((((...(.((......)).)..)))))))))((((((((.--...))))).)))))))))))))((((....)))))))))))).... ( -50.70) >consensus UGCGAAUACCGAGACUCUGGCUGAAACACCCAGACAAGGGCGAAGAAUCGGCGAGUCUGGGCUGUAGUACGUAUGUAUGUAUGUGAGGUGGAUUCGGCACAAGUGUGCUAUGUACAGCUC ........((.((((((..(((((..(((((......))))).....)))))))))))))((((((((((....)))).................(((((....)))))...)))))).. (-30.51 = -31.45 + 0.94)

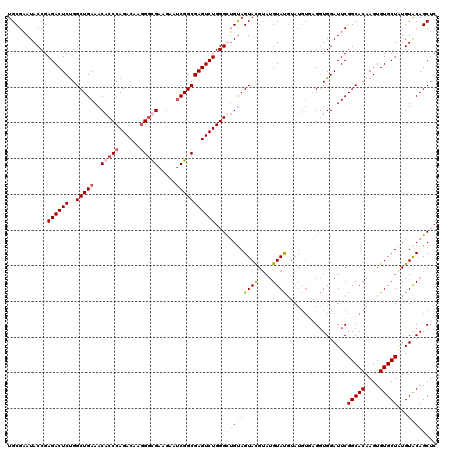
| Location | 847,661 – 847,773 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -28.91 |
| Energy contribution | -27.85 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
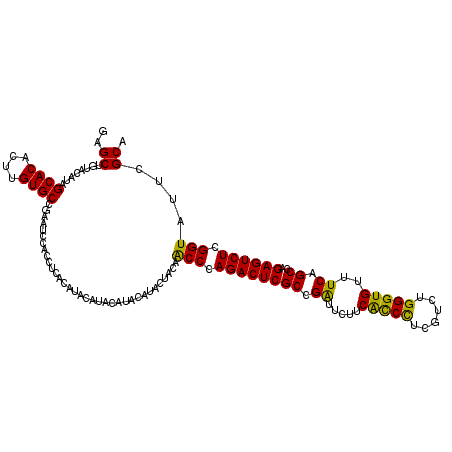
>3R_DroMel_CAF1 847661 112 - 27905053 GAGCUGUACAUAGCACACUUGUGCCGAAUCCACC--------AUACAUACGUACUACAACCCAGACUCGCCGAUUCUUCGCCCUCGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCA ..((.((((...((((....)))).(....)...--------........))))....(((.((((((((.((.....(((((......)))))..)).))..)))))).)))....)). ( -32.10) >DroSec_CAF1 23331 120 - 1 GAGCUGUACACAGCACACUUGUGCCGAAUCCACCUCACAUACAUACAUACAUACUGCAGCCCAGACUCGCCGAUUCUUCACCCUCGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCA (((.((....(.((((....)))).)....)).)))...................((.(((.((((((((.((.....(((((......)))))..)).))..)))))).)))....)). ( -32.50) >DroSim_CAF1 22579 120 - 1 GAGCUGUACAUAGCACACUUGUGCCGAAUCCACCUCACAUACAUACAUACAUACUACAGCCCAGACUCGCCGAUUCUUCACCCUUGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCA ..((((((....((((....)))).((.......))..................))))))((((((((((.((.....(((((......)))))..)).))..)))))).))........ ( -32.30) >DroEre_CAF1 24381 118 - 1 GAGCUGCACAUAGCACACUUGUGCCGAAUCCAGCGUACAUACGUAC--UCGUACUACCACCCAGACUCGCCGGCUCCUCGUCUUUGUCUGGGUGUUUCAGCCGGAGUCUCGGUAUGCGCA .....((.((((((((....))))(((.(((.((((((....))))--.........(((((((((.....(((.....)))...))))))))).....)).)))...))).)))).)). ( -41.10) >consensus GAGCUGUACAUAGCACACUUGUGCCGAAUCCACCUCACAUACAUACAUACAUACUACAACCCAGACUCGCCGAUUCUUCACCCUCGUCUGGGUGUUUCAGCCAGAGUCUCGGUAUUCGCA ..((........((((....))))..................................(((.((((((((.((.....(((((......)))))..)).))..)))))).)))....)). (-28.91 = -27.85 + -1.06)

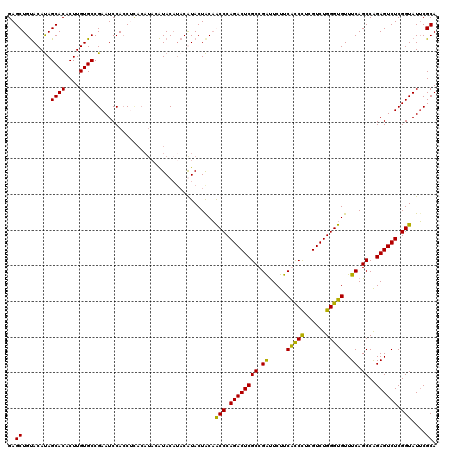
| Location | 847,739 – 847,837 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -26.34 |
| Energy contribution | -27.90 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
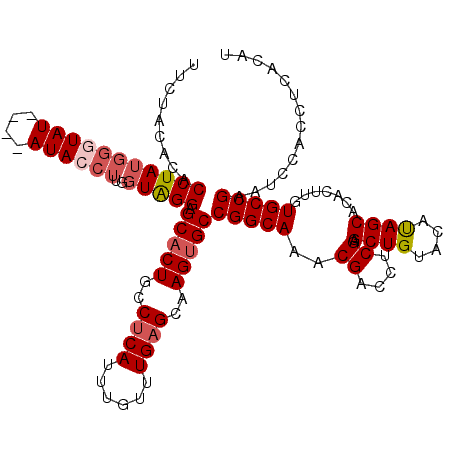
>3R_DroMel_CAF1 847739 98 - 27905053 UUCUACACACCUAUGGG----------------UAGGAGCACUGCCUCAUUUGUUUGAGCAAGUGCCGGCAAACGACCUCGAGCUGUACAUAGCACACUUGUGCCGAAUCCACC------ ........(((....))----------------).((((((((..((((......))))..)))))(((((..((....)).((((....)))).......)))))..)))...------ ( -29.20) >DroSec_CAF1 23411 120 - 1 UUCUACACACCUAUGGGUAUACAUAUAACUUGGUAGGAGCACUGCCUCAUUUGUUUGAGCAAGUGCCGGCAAACGACCUCGAGCUGUACACAGCACACUUGUGCCGAAUCCACCUCACAU ........(((....))).............(((.((((((((..((((......))))..)))))(((((..((....)).((((....)))).......)))))..))))))...... ( -35.00) >DroSim_CAF1 22659 120 - 1 UUCUACACACCUAUGGGUAUACAUAUACCUUGGUAGGAGCACUGCCUCAUUUGUUUGAGCAAGUGCCGGCAAACGACCUCGAGCUGUACAUAGCACACUUGUGCCGAAUCCACCUCACAU ........(((...((((((....)))))).))).((((((((..((((......))))..)))))(((((..((....)).((((....)))).......)))))..)))......... ( -35.90) >DroEre_CAF1 24459 116 - 1 UUCCCCAUACCUAUGGGUAU----AUACCUUGGUGGGUGCUCUGCCCCAUUUGUUUGAGCAAGUGCCGGCAAACGACCUCGAGCUGCACAUAGCACACUUGUGCCGAAUCCAGCGUACAU .......((((...((((..----..)))).)))).((((..((((.(((((((....)))))))..))))..((....)).((((......((((....))))......)))))))).. ( -34.00) >consensus UUCUACACACCUAUGGGUAU____AUACCUUGGUAGGAGCACUGCCUCAUUUGUUUGAGCAAGUGCCGGCAAACGACCUCGAGCUGUACAUAGCACACUUGUGCCGAAUCCACCUCACAU .........(((((((((((....))))))..))))).(((((..((((......))))..)))))(((((..((....)).((((....)))).......))))).............. (-26.34 = -27.90 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:41 2006