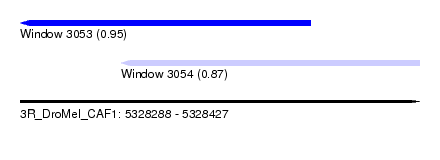
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,328,288 – 5,328,427 |
| Length | 139 |
| Max. P | 0.951797 |

| Location | 5,328,288 – 5,328,389 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.96 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5328288 101 - 27905053 UGCUCCACCGCGCUCUCUUCGUUCGCUCUUCGCGUAUCUGCGAGCUACUCGAACU--------------CUGCGAGCUAAACGAAUCGGAAUCGCCAAAGGUU-----CAGUUGUUCGAC .((......))((((.(...(((((...((((((....)))))).....))))).--------------..).))))....(((((..(((((......))))-----)....))))).. ( -27.10) >DroSim_CAF1 33826 91 - 1 --------CGCGCUCUCCUCGCUCGCUCUUCGCGUAUCUGCGAGAUACUCGAAC----------------UGCGAGUGAAACGAAUCCGAAUCGCCAACGGUU-----CAGUUGUUCGAC --------..........((((((((..((((.(((((.....))))).)))).----------------.))))))))..((((((.((((((....)))))-----).)..))))).. ( -34.00) >DroYak_CAF1 33635 111 - 1 -------CUGCG--CUCUUCGCUCGCUCUUCGCGUAUCUGCGAGAUACUCGUACUGCCGGAUACUCGAACUGCGAGCGAAACAGAAACAACUCGCCGAUGGUUCACUUCAGUUGUUUGAC -------(((..--...(((((((((..((((.(((((((((((...)))))......)))))).))))..))))))))).)))((((((((....((....)).....))))))))... ( -38.40) >consensus _______CCGCGCUCUCUUCGCUCGCUCUUCGCGUAUCUGCGAGAUACUCGAACU______________CUGCGAGCGAAACGAAUCCGAAUCGCCAAAGGUU_____CAGUUGUUCGAC ..................((((((((..((((.(((((.....))))).))))..................))))))))(((((.........(((...))).........))))).... (-18.47 = -18.59 + 0.12)



| Location | 5,328,323 – 5,328,427 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.25 |
| Mean single sequence MFE | -33.94 |
| Consensus MFE | -19.36 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5328323 104 - 27905053 UAAGGAAACAUUGUGU--GCCAAGUGCUAAGAGAGAGAGCUGCUCCACCGCGCUCUCUUCGUUCGCUCUUCGCGUAUCUGCGAGCUACUCGAACU--------------CUGCGAGCUAA ...(....)...((.(--((..(((.....((((((((((.((......)))))))))..((((((.............))))))..)))..)))--------------..))).))... ( -31.02) >DroSim_CAF1 33861 91 - 1 UAAGGAAACAUUGUGU--GCCAACUGCUAAGAGAGAG-----------CGCGCUCUCCUCGCUCGCUCUUCGCGUAUCUGCGAGAUACUCGAAC----------------UGCGAGUGAA ...(....)...((((--((...((......))...)-----------))))).....((((((((..((((.(((((.....))))).)))).----------------.)))))))). ( -34.40) >DroYak_CAF1 33675 108 - 1 UAAGGAAAUAUUAUAUUGGCCAACUGCUAAGAGAGAA----------CUGCG--CUCUUCGCUCGCUCUUCGCGUAUCUGCGAGAUACUCGUACUGCCGGAUACUCGAACUGCGAGCGAA ...............(((((.....))))).((((..----------.....--))))((((((((..((((.(((((((((((...)))))......)))))).))))..)))))))). ( -36.40) >consensus UAAGGAAACAUUGUGU__GCCAACUGCUAAGAGAGAG__________CCGCGCUCUCUUCGCUCGCUCUUCGCGUAUCUGCGAGAUACUCGAACU______________CUGCGAGCGAA ...((..((.....))...)).........(((((.................))))).((((((((..((((.(((((.....))))).))))..................)))))))). (-19.36 = -20.15 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:14 2006