| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,298,912 – 5,299,139 |
| Length | 227 |
| Max. P | 0.940168 |

| Location | 5,298,912 – 5,299,023 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.40 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -20.45 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5298912 111 + 27905053 --GUUCAGUGGCUCAGUGGAUUUGUUGGUCAAGAGAUCAACAGAUCAACAGAUCAACAGAUCAAUAGAGCACUAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCU --....((((((...((.((((((((((((....)))))))))))).)).((((....))))...((((((((....)))))...((((((......))))))))).)))))) ( -38.70) >DroSec_CAF1 3552 103 + 1 --GUUCAGUGGCUCAGCGGAUUUGUUGGUCAAGAAAUCAACAGA--------UCAACAGAUCAAUAGAGCACCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCU --....((((((...((.((((((((((((............))--------))))))))))......((((......)))))).((((((......))))))....)))))) ( -33.10) >DroSim_CAF1 3146 103 + 1 --GUUCAGUGGCUCAGUGGAUUUGUUGGUCAAGAGAUCAACGGA--------UCAACAGAUCAAUAGAGCACCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCU --....((((((...((.((((((((((((....))))))))))--------)).))(((((......((((......)))).........(((....)))))))).)))))) ( -34.80) >DroEre_CAF1 3414 97 + 1 UCGCUCGGUGGCUCGGUGGUUUUGUGGGU----------------CAAGGGAUCAAGAGAUCAAUGGAGCACUAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCU ......((((((..(((...(((((((((----------------.((..((((....))))...((.(((((....))))).))...)).)))))))))..)))..)))))) ( -31.50) >DroYak_CAF1 3156 111 + 1 --GUUAAGUGGCUCAGUGGUUUUGUGGAUCAUGAGAUCAUGAGAUCAAGAGAUCAAGAGAUCAAUAGACCCCCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCU --....((((((...(((((((..((...))..)))))))(((((((...((((....))))...........(((.((((.....)))).)))......))))))))))))) ( -29.60) >consensus __GUUCAGUGGCUCAGUGGAUUUGUUGGUCAAGAGAUCAACAGA_CAA__GAUCAACAGAUCAAUAGAGCACCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCU ......((((((.....((.((((((((((............................))))))))))((((......)))).))((((((......))))))....)))))) (-20.45 = -20.97 + 0.52)

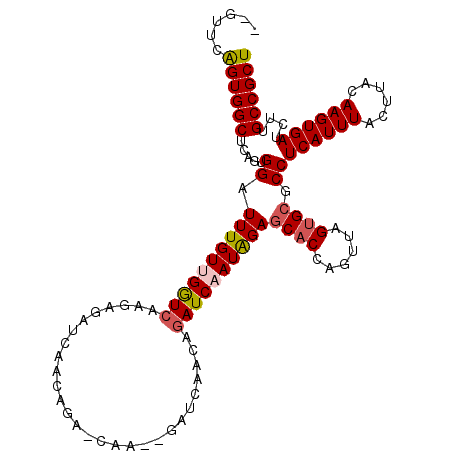

| Location | 5,298,946 – 5,299,063 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.57 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -27.24 |
| Energy contribution | -28.24 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5298946 117 + 27905053 UCAACAGAUCAACAGAUCAACAGAUCAAUAGAGCACUAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCUUGAAUGCCUUCGAGCAGAGCGCACAGAUGUAUUUCCGCUC ......((((....))))............((((........((((((.((.........((((....))))..(((((((....))))))).)))))))).((......)).)))) ( -31.80) >DroSec_CAF1 3586 109 + 1 UCAACAGA--------UCAACAGAUCAAUAGAGCACCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCUUGAAUGCCUUCGAGCAGAGCGCACAGAUGUAUUUCCGCUC ......((--------((....))))....((((........((((((.((.........((((....))))..(((((((....))))))).)))))))).((......)).)))) ( -31.80) >DroSim_CAF1 3180 109 + 1 UCAACGGA--------UCAACAGAUCAAUAGAGCACCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCUUGAAUGCUUUCGAGCAGAGCGCACAGAUGUAUUUCCGCUC ......((--------((....))))....((((........((((((.((.........((((....))))..(((((((....))))))).)))))))).((......)).)))) ( -32.40) >DroEre_CAF1 3443 108 + 1 ---------CAAGGGAUCAAGAGAUCAAUGGAGCACUAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCUUGAAUGCCUUCGAGCAGAGCGCACAGAUGUAUUUCCGCUC ---------.....((((....))))...((((.....(((.((((((.((.........((((....))))..(((((((....))))))).)))))))).)))....)))).... ( -31.50) >DroYak_CAF1 3190 117 + 1 UCAUGAGAUCAAGAGAUCAAGAGAUCAAUAGACCCCCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCUUGAAUGCCUUCGAGCAGAGCGCACAGAUGUAUUUCCGCUC ....(((((((...((((....))))................((((((.((.........((((....))))..(((((((....))))))).))))))))...)).)))))..... ( -30.10) >consensus UCAACAGA_CAA__GAUCAACAGAUCAAUAGAGCACCAGUUAGUGCGCCUCAUUUACUUACAAGUGAUCUUGCCGCUUGAAUGCCUUCGAGCAGAGCGCACAGAUGUAUUUCCGCUC ..............((((....))))....((((........((((((.((.........((((....))))..(((((((....))))))).)))))))).((......)).)))) (-27.24 = -28.24 + 1.00)

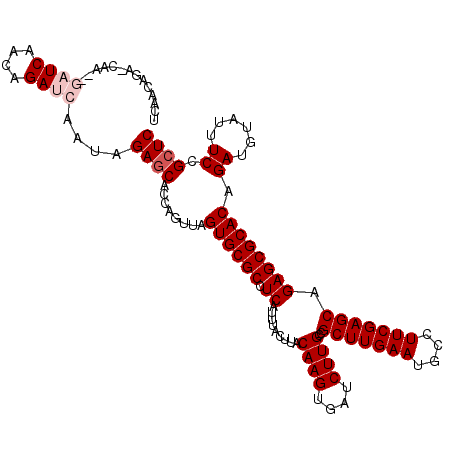

| Location | 5,299,023 – 5,299,139 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.70 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.22 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5299023 116 - 27905053 ----GGACUUUUUAUAGAUUUGUUGCAAGAUCUUUGGAAAUGCCUCGGCGCUGGCCAAGCACAAACUGACGCACAGCGACGAGCGGAAAUACAUCUGUGCGCUCUGCUCGAAGGCAUUCA ----(((.((((((.((((((......)))))).))))))(((((...(((((((..((......))...)).))))).((((((((..(((....)))...)))))))).)))))))). ( -37.20) >DroVir_CAF1 3453 116 - 1 ----GUACUUUUUAUAGAUUUGUAGCAAGAUCUUUGGAAAUGCUUCGGCACUGGCCAAGCACAAACUGACGCACAGCGACGAGCGGAAAUAUAUUUGUGCGCUCUGCUCGAAGGCAUUCA ----....((((((.((((((......)))))).))))))(((....)))...(((.............(((...))).((((((((.(((....)))....))))))))..)))..... ( -31.30) >DroGri_CAF1 4384 116 - 1 ----AUACUUUUUAUAGAUUUGUAGCAAGAUAUUCGGAAAUGCCUCGGCGUUGGCCAAGCACAAAUUGACGCACAGCGACGAACGGAAAUAUAUUUGUGCGCUCUGCUCGAAGGCAUUCA ----...((((.....(((((((.((..(.....)((.(((((....)))))..))..)))))))))((.(((.((((....(((((......))))).)))).)))))))))....... ( -28.90) >DroWil_CAF1 3183 114 - 1 ------CCUUUUUGUAGAUUUGCUGCAAAAUCUUUGGAAAUGCUUCGGCGUUGGCCAAGCACAAACUGACGCACAGCGACGAACGGAAAUACAUUUGCGCGCUUUGCUCUAAGGCAUUCA ------...((((((((.....)))))))).(((((((..(((((.(((....)))))))).........(((.((((.((((.(......).))))..)))).))))))))))...... ( -32.70) >DroMoj_CAF1 3530 111 - 1 ---------UUUAAUAGAUUUGCAGCAAGAUCUUUGGAAAUGCCUCGGCACUAGCCAAGCACAAACUGACGCACAGCGACGAACGGAAAUACAUCUGUGCGCUCUGCUCGAAGGCAUUCA ---------(((((.((((((......)))))))))))(((((((((((....)))...........((.(((.((((....(((((......))))).)))).)))))).))))))).. ( -33.30) >DroAna_CAF1 3723 120 - 1 UUCUGGCCUUUUUAUAGAUUUGUUGCAAGAUCUUUGGAAAUGCCUCGGCACUGGCCAAGCACAAACUGACGCACAGCGACGAACGGAAAUACAUCUGUGCGCUCUGCUCGAAGGCAUUCA ...(((((((((((.((((((......)))))).))))))(((....)))..))))).((......(((.(((.((((....(((((......))))).)))).))))))...))..... ( -35.80) >consensus ____G_ACUUUUUAUAGAUUUGUAGCAAGAUCUUUGGAAAUGCCUCGGCACUGGCCAAGCACAAACUGACGCACAGCGACGAACGGAAAUACAUCUGUGCGCUCUGCUCGAAGGCAUUCA ..........((((.((((((......)))))).))))(((((((.(((....)))...........((.(((.((((....(((((......))))).)))).)))))..))))))).. (-29.44 = -29.22 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:21:00 2006