
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,283,463 – 5,283,559 |
| Length | 96 |
| Max. P | 0.821245 |

| Location | 5,283,463 – 5,283,559 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -16.02 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5283463 96 + 27905053 -----------UUAUGUU--AUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUUAAAUGCUGUUAGCAAUUUCU-UAAAUCACAUCCGACUGUUUCUGCUUAAUGGAAUCC-------- -----------...((((--(...(((((((((.((((((....))))))...))))))))))))))......-.........((((...((....))....))))....-------- ( -18.50) >DroVir_CAF1 1941 100 + 1 -----------UU--UUU--AUCGAUAGUAUUUUUAUGCAUUUAUGCAUAGUUAAAUGCUGUUAGCAAUUUUU-UAAAUCACAUCCGACUGUUUCU-CUCAAUCAAAUCUC-GCUUCU -----------..--...--...(((((((((((((((((....)))))))..))))))))))..........-............((.((.....-..)).)).......-...... ( -14.60) >DroGri_CAF1 3652 99 + 1 -----------UU-UUUU--AUCGAUAGUAUUUUUAUGCAUUUAUGCAUAGUUAAAUGCUGUUAGCAAUUUUU-UAAAUCACAUCCGACUGUAUCU-AUCAAUGGAAUUUCCG---CC -----------..-....--...(((((((((((((((((....)))))))..)))))))))).((.......-......(((......)))....-......((.....)))---). ( -17.20) >DroWil_CAF1 3241 109 + 1 UCAUUUGUAAUUCAUGUUAAACCGAUAGUAUUUUUAUGCAUUUAUGCAUAGUUAAAUGCUGUUAGCAAAUUUU-UUAAUCACAUCCGACUGUUUCUGCCUAAUCAAAUCU-------- ..(((((((((....))).....(((((((((((((((((....)))))))..)))))))))).))))))...-....................................-------- ( -15.40) >DroMoj_CAF1 1441 88 + 1 -----------UU-UUUU--AUCGAUAGUAUUUUUAUGCAUUUAUGCAUAGUUAAAUGCUGUUAGCAAUUUUU-UGAAUCACAUCCGACUGUUUCU-CUCC-----------G---CC -----------..-....--...(((((((((((((((((....)))))))..))))))))))..........-.(((.((........)).))).-....-----------.---.. ( -14.40) >DroAna_CAF1 1485 97 + 1 -----------UUAUACU--AUCGAUAGUAUUUUUAUGCAUUUAUGCAUAUUUAAAUGCUGUUAGCAAUUUCUAUAAAUCACAUCCGACUGUUUCUUCUUAAUGGAAUCC-------- -----------.......--...((((((((((.((((((....))))))...))))))))))...........................(((((........)))))..-------- ( -16.00) >consensus ___________UU_UUUU__AUCGAUAGUAUUUUUAUGCAUUUAUGCAUAGUUAAAUGCUGUUAGCAAUUUUU_UAAAUCACAUCCGACUGUUUCU_CUCAAUGGAAUCU________ .......................((((((((((.((((((....))))))...))))))))))....................................................... (-14.23 = -14.23 + -0.00)



| Location | 5,283,463 – 5,283,559 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -15.40 |
| Consensus MFE | -11.12 |
| Energy contribution | -10.98 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
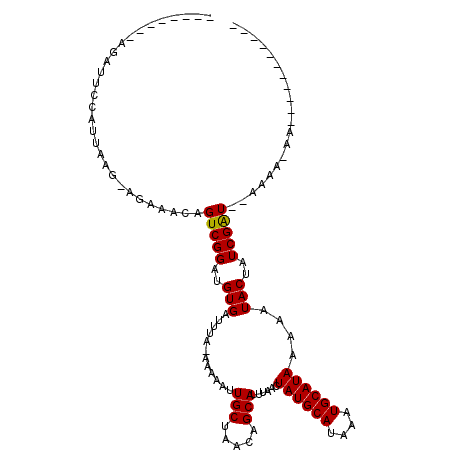
>3R_DroMel_CAF1 5283463 96 - 27905053 --------GGAUUCCAUUAAGCAGAAACAGUCGGAUGUGAUUUA-AGAAAUUGCUAACAGCAUUUAAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAU--AACAUAA----------- --------((((..((((..((.......))..))))..)))).-.((...(((.....)))......((((((....)))))).........))...--.......----------- ( -12.80) >DroVir_CAF1 1941 100 - 1 AGAAGC-GAGAUUUGAUUGAG-AGAAACAGUCGGAUGUGAUUUA-AAAAAUUGCUAACAGCAUUUAACUAUGCAUAAAUGCAUAAAAAUACUAUCGAU--AAA--AA----------- .((.((-...(((((((((..-.....)))))))))(..(((..-...)))..).....)).......((((((....)))))).........))...--...--..----------- ( -16.40) >DroGri_CAF1 3652 99 - 1 GG---CGGAAAUUCCAUUGAU-AGAUACAGUCGGAUGUGAUUUA-AAAAAUUGCUAACAGCAUUUAACUAUGCAUAAAUGCAUAAAAAUACUAUCGAU--AAAA-AA----------- ..---.((.....))((((((-((.....((..(((((...(((-.........)))..)))))..))((((((....))))))......))))))))--....-..----------- ( -19.90) >DroWil_CAF1 3241 109 - 1 --------AGAUUUGAUUAGGCAGAAACAGUCGGAUGUGAUUAA-AAAAUUUGCUAACAGCAUUUAACUAUGCAUAAAUGCAUAAAAAUACUAUCGGUUUAACAUGAAUUACAAAUGA --------.(((((..((((((.((....((..(((((..(((.-.........)))..)))))..))((((((....)))))).........)).))))))...)))))........ ( -16.10) >DroMoj_CAF1 1441 88 - 1 GG---C-----------GGAG-AGAAACAGUCGGAUGUGAUUCA-AAAAAUUGCUAACAGCAUUUAACUAUGCAUAAAUGCAUAAAAAUACUAUCGAU--AAAA-AA----------- ..---(-----------((((-.......((..(((((....((-(....)))......)))))..))((((((....))))))......)).)))..--....-..----------- ( -12.60) >DroAna_CAF1 1485 97 - 1 --------GGAUUCCAUUAAGAAGAAACAGUCGGAUGUGAUUUAUAGAAAUUGCUAACAGCAUUUAAAUAUGCAUAAAUGCAUAAAAAUACUAUCGAU--AGUAUAA----------- --------..................((.(((((..(..((((....))))..).....(((((((........)))))))............)))))--.))....----------- ( -14.60) >consensus ________AGAUUCCAUUAAG_AGAAACAGUCGGAUGUGAUUUA_AAAAAUUGCUAACAGCAUUUAACUAUGCAUAAAUGCAUAAAAAUACUAUCGAU__AAAA_AA___________ .............................(((((..(((............(((.....)))......((((((....))))))....)))..))))).................... (-11.12 = -10.98 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:53 2006