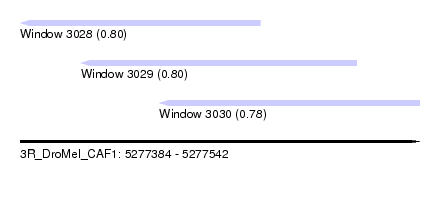
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,277,384 – 5,277,542 |
| Length | 158 |
| Max. P | 0.804247 |

| Location | 5,277,384 – 5,277,479 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5277384 95 - 27905053 UGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACACCCGAGUAGCUCCAUUUUCCCAUUUUC--------CCAUCUUC---CCUUCGUCACGCCCCCUCAGCC (((((((((((..(....)..))))))((((........))))............))))).....--------........---...................... ( -18.30) >DroSim_CAF1 45235 98 - 1 UGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACAC-CGAGUAACUCCAUUUUCCCAUUUUCC-------CCAUCUUCCCCCCUUCGUCACGCCCCCUCAGCC (((((((((((..(....)..))))))(((((.....))-)))............)))))......-------................................. ( -19.60) >DroYak_CAF1 51909 103 - 1 UGGGAUCGGAUCUGAUCUCACAUCCGAUCGGUUCGAUCCCCGAGUAUCUCCAAUUCCCCAACUUCCCAUCUUUCCAUCUUC---CCUUCGUCACGCCCCCUCAGCC .(((((((((.((((((........))))))))))))))).((((.......)))).........................---...................... ( -22.20) >consensus UGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACACCCGAGUAACUCCAUUUUCCCAUUUUCC_______CCAUCUUC___CCUUCGUCACGCCCCCUCAGCC (((((((((((..(....)..))))))))((....((......))....))......))).............................................. (-16.32 = -16.10 + -0.22)

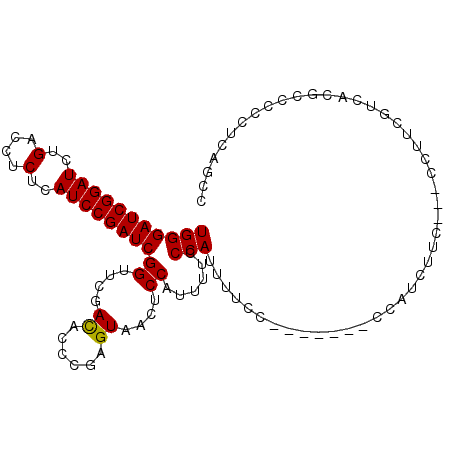
| Location | 5,277,408 – 5,277,517 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -30.43 |
| Energy contribution | -32.10 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5277408 109 - 27905053 ACGUCGAAAGCGGGACGCGGGUGGGUCCUUUGAACCGAUGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACACCCGAGUAGCUCCAUUUUCCCAUUUUC--------CCAUCU ..(.(....))((((...(((.((((...((((((((((.((((.(((.......))).)))).)))))))))).))))(((...)))......)))....))--------)).... ( -40.20) >DroSim_CAF1 45262 109 - 1 ACGUCGAAAGCGGGACGCGGGUGGGUCCUUUGAACCGAUGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACAC-CGAGUAACUCCAUUUUCCCAUUUUCC-------CCAUCU ..(..(((((.((((...((((((((...((((((((((.((((.(((.......))).)))).)))))))))).))-)(((...)))))))))))).))))).-------.).... ( -37.10) >DroYak_CAF1 51933 113 - 1 ACGACGAAAGCGGGACGCGG----GUCCUUUGAACCGAUGGGAUCGGAUCUGAUCUCACAUCCGAUCGGUUCGAUCCCCGAGUAUCUCCAAUUCCCCAACUUCCCAUCUUUCCAUCU .....(((((.((((.(.((----(......((((((..(((((((((.((((((........))))))))))))))))).)).)).......)))...).))))..)))))..... ( -37.22) >consensus ACGUCGAAAGCGGGACGCGGGUGGGUCCUUUGAACCGAUGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACACCCGAGUAACUCCAUUUUCCCAUUUUCC_______CCAUCU .((.(....)))(((..((((((......((((((((((.((((.(((.......))).)))).))))))))))))))))......)))............................ (-30.43 = -32.10 + 1.67)

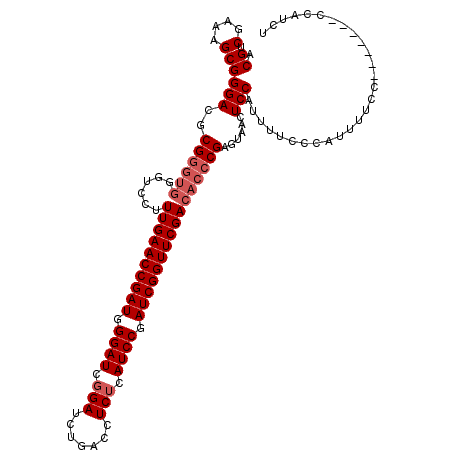
| Location | 5,277,439 – 5,277,542 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -21.54 |
| Energy contribution | -23.10 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5277439 103 - 27905053 CAAUAAAAUCCAAACAAGCAUAAACACGUCGAAAGCGGGACGCGGGUGGGUCCUUUGAACCGAUGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACACC ...........................(((......(((((.(....).)))))..((((((((.((((.(((.......))).)))).)))))))))))... ( -32.50) >DroSim_CAF1 45294 102 - 1 CAAUAAAAUCCAAACAAGCAUAAACACGUCGAAAGCGGGACGCGGGUGGGUCCUUUGAACCGAUGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACAC- ...........................(((......(((((.(....).)))))..((((((((.((((.(((.......))).)))).)))))))))))..- ( -32.50) >DroYak_CAF1 51972 99 - 1 CAAUAAAAUCCAAACAAGCAUAAACACGACGAAAGCGGGACGCGG----GUCCUUUGAACCGAUGGGAUCGGAUCUGAUCUCACAUCCGAUCGGUUCGAUCCC ..........((((............((.(....)))((((....----)))))))).......(((((((((.((((((........))))))))))))))) ( -30.60) >DroAna_CAF1 48793 93 - 1 CAAUAAAAUCCAAACAAGCAUAAACACGUCGAAAGCGGGUCGCAG----GACAUCCGAACCGGAUGGAUAGGAUGUGACAUUGCGCCCCAUAUCUUC------ ...........................(.(....))(((.(((((----.((((((...((....))...))))))....))))).)))........------ ( -23.30) >consensus CAAUAAAAUCCAAACAAGCAUAAACACGUCGAAAGCGGGACGCGG____GUCCUUUGAACCGAUGGGAUCGGAUCUGACCUCUCAUCCGAUCGGUUCGACAC_ ..........................((.(....)))((((........)))).((((((((((.((((.(((.......))).)))).)))))))))).... (-21.54 = -23.10 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:51 2006