
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,273,350 – 5,273,509 |
| Length | 159 |
| Max. P | 0.976605 |

| Location | 5,273,350 – 5,273,469 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -28.33 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.780035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5273350 119 - 27905053 CUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUUGCGCCAGAAACAUGUUGAAUUUCAACUAA-ACUACGACCGCUCGGAC ((((.(.((..(((((.......(.(((((.((((...((((((((...((......))..))))))))....)))).))))))..((((.....))))..)-))))..)).).)))).. ( -33.50) >DroVir_CAF1 46290 119 - 1 CUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUCGCGCCAGAGACAUGUUGAAUUUCAACUAA-ACUACGACCGUUCGGAC ((((.(.((..(((((.......(.(((((.((((...((((((((...((......))..))))))))....)))).))))))..((((.....))))..)-))))..)).).)))).. ( -33.60) >DroGri_CAF1 56717 119 - 1 CUGACCAGUAAUGGUAACAAUGCGAUUUCUAGUGCCAGGUGCGAAAAUGCCCUUUUUGGAAAUUUGCACAUUAGCGCCAGAAACAUGUUGAAUUUCAACUAA-ACUACGACCGUUCGGAC (((((((....))))......(((.(((((.((((...(((((((....((......))...)))))))....)))).)))))...((((.....))))...-........))).))).. ( -29.70) >DroWil_CAF1 58012 119 - 1 GUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAAAUGCCCUUUUUCGAAAUUUGCACAUUUGCGCCAGAAACAUGUUGAAUUUCAACUAA-ACUACGACCAUUUGGAC (((((((....)))))))....((.(((((.((((...(((((((.................)))))))....)))).)))))...((((.....))))...-....)).((....)).. ( -29.73) >DroMoj_CAF1 48024 119 - 1 CUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCUUUUUUGGAAAUUUGCACAUUUGCGCCAGAAGCAUGUUGAAUUUCAACUAA-ACUACGACCGUUCGGAC ((((.(.((..(((((...((((...((((.((((...((((((((...((......))..))))))))....)))).))))))))((((.....))))..)-))))..)).).)))).. ( -36.80) >DroAna_CAF1 44531 120 - 1 CUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUUGCGCCAGAAACAUGUAGAAUUCCAAAAAACACUACGCCCGUUGGGGC .((((((....))))))......(.(((((.((((...((((((((...((......))..))))))))....)))).))))))..((((..............))))((((....)))) ( -33.84) >consensus CUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUUGCGCCAGAAACAUGUUGAAUUUCAACUAA_ACUACGACCGUUCGGAC .((((((....))))))((((..(.(((((.((((...((((((((...((......))..))))))))....)))).))))))..))))....................((....)).. (-28.33 = -28.88 + 0.56)
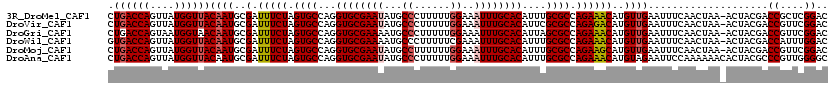


| Location | 5,273,389 – 5,273,509 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -33.87 |
| Energy contribution | -34.40 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5273389 120 - 27905053 UCUGUUCAAUUAGAUAUUGCGCUUUGGCAAUGGCACUCGGCUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUUGCGCCAG ........((((((.((((((((..(((....)).)..)))((((((....))))))....))))).))))))....(((((((((.(((((.....))......))).))))))))).. ( -37.40) >DroPse_CAF1 50136 120 - 1 UCUGCUCAAUUAGAAACUGCGCUUUGGCAAUGGCGUUCGGCUGACCAGUAAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUUGCGCCAG ((((......))))..((((((.((((((.((((((...(.((((((....)))))))...)))....))).))))))((((((((...((......))..))))))))....))).))) ( -37.30) >DroGri_CAF1 56756 120 - 1 UCUGUUCAAUUAGAAACUGCGCCUUGGCAAUGGCAUUCGGCUGACCAGUAAUGGUAACAAUGCGAUUUCUAGUGCCAGGUGCGAAAAUGCCCUUUUUGGAAAUUUGCACAUUAGCGCCAG ((((......))))..((((((.((((((((.(((((.(....((((....))))..)))))).))......))))))(((((((....((......))...)))))))....))).))) ( -34.90) >DroWil_CAF1 58051 120 - 1 UCUGUUCAAUUAGACAUUGCGCUUUGGCAAUGGCACUCGGGUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAAAUGCCCUUUUUCGAAAUUUGCACAUUUGCGCCAG ((((......))))....((((..(((((((((((((((.(((((((....)))))))....)))......))))))....((((((......))))))....)))).))...))))... ( -36.80) >DroMoj_CAF1 48063 120 - 1 UUUGUUCAAUUAGAUACUGCGCUUUGGCAAUGGCACUCGGCUGACCAGUUAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCUUUUUUGGAAAUUUGCACAUUUGCGCCAG ................((((((.((((((.(((...(((..((((((....)))))).....)))...))).))))))((((((((...((......))..))))))))....))).))) ( -35.30) >DroPer_CAF1 50679 120 - 1 UCUGCUCAAUUAGAAACUGCGCUUUGGCAAUGGCGUUCGGCUGACCAGUAAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUUGCGCCAG ((((......))))..((((((.((((((.((((((...(.((((((....)))))))...)))....))).))))))((((((((...((......))..))))))))....))).))) ( -37.30) >consensus UCUGUUCAAUUAGAAACUGCGCUUUGGCAAUGGCACUCGGCUGACCAGUAAUGGUUACAAUGCGAUUUCUAGUGCCAGGUGCGAAUAUGCCCUUUUUGGAAAUUUGCACAUUUGCGCCAG ((((......))))....((((.((((((.(((...(((..((((((....)))))).....)))...))).))))))((((((((...((......))..))))))))....))))... (-33.87 = -34.40 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:49 2006