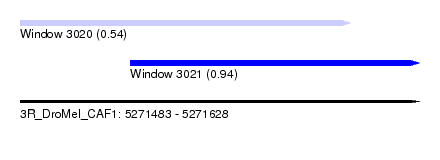
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,271,483 – 5,271,628 |
| Length | 145 |
| Max. P | 0.943254 |

| Location | 5,271,483 – 5,271,603 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -42.86 |
| Consensus MFE | -19.79 |
| Energy contribution | -20.27 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5271483 120 + 27905053 CCACGAUCGGCGGUGCAAACUCGGCGGCCAGCCUGCUAGUCAACGGCACCGGCAUCAAUUUGUCCAUCAAUCUGGGCUCACCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACACAGCUGC .........(((((((.......))(((.((((.(((((.....(((.((((.........(((((......))))).........)))).)))....)))))))))))).....))))) ( -42.57) >DroVir_CAF1 44732 120 + 1 CGGCCAUCGGCGGUGCGGGUACUGCAACCAGUCUGUUGGUCAAUGGCGCAGGCAUUAAUCUAUCCCUCAACCUGGGCUCACCUAACCCAGCGCCCAAUCUGGCAGUUGCCACACAAUUGC .(((((.(((((((((((...)))).))).).))).)))))...((((((((.((......)).))).....((((.........))))))))).......(((((((.....))))))) ( -38.20) >DroGri_CAF1 54967 120 + 1 CUGCAAUCGGUGGUGCGGCAACCGCAGCCAGUCUUUUGGUCAAUGGCACCGGCAUCAACUUAUCCCUCAAUUUGGGCGCGCCAAAUCCGGCGCCAAAUCUAGCUGUUGCCAAUCAGCUGC ..(((...(.((((..((((((.((.(((((....)))))...((((.((((...........(((.......)))..........)))).))))......)).)))))).)))).)))) ( -40.20) >DroWil_CAF1 55779 120 + 1 CAUCGAUUGGCGGCGCUGGCUCCGCAUCCAGUGUCCUAGUCAAUGGUACUGGCAUCAAUUUGUCCCUCAAUCUGGGUGCUCCGAAUCCUGCGCCCAAUCUAGCAGUUGCCACACAAUUGC (((.((((((.((((((((........)))))))))))))).))).....((((......))))........(((((((..........))))))).....(((((((.....))))))) ( -41.40) >DroMoj_CAF1 46290 120 + 1 CGGCCAUCGGCGGCGCCGGCACUGCGGCCAGCCUCCUGGUCAGCAAUGCGGGCAUCAACUUGUCCCUCAACUUGAGCGCCCCCAAUCCGACGCCCAACCUGGCGUUUGCCACACAGCUGU .(((....((.((((((.(((.(((((((((....)))))).))).))).))).((((.(((.....))).))))..))).)).....((((((......)))))).))).......... ( -49.50) >DroAna_CAF1 42620 120 + 1 CAACGAUCGGCGGCACCAGCUCCGCUGCCAGCCUGCUAGUCAACGGCACCGGCAUCAACUUGUCUAUCAACCUGGGCGCCCCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACUCAACUCC ....(((..(((((..((((...))))...))).))..)))...((((((((((......))))........((((((((........)))))))).......)).)))).......... ( -45.30) >consensus CAGCGAUCGGCGGCGCCGGCACCGCAGCCAGCCUGCUAGUCAACGGCACCGGCAUCAACUUGUCCCUCAACCUGGGCGCACCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACACAACUGC .........((((........))))...((((......(.((((.((...((((......))))........(((((((..........))))))).....)).)))).).....)))). (-19.79 = -20.27 + 0.48)


| Location | 5,271,523 – 5,271,628 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5271523 105 + 27905053 CAACGGCACCGGCAUCAAUUUGUCCAUCAAUCUGGGCUCACCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACACAGCUGCUCGAGCAUAUUAAGGUUAG---ACUUG-----G ....(((.((((.........(((((......))))).........)))).)))...((.((((((((.....)))))))).))......((((.....---.))))-----. ( -32.17) >DroVir_CAF1 44772 106 + 1 CAAUGGCGCAGGCAUUAAUCUAUCCCUCAACCUGGGCUCACCUAACCCAGCGCCCAAUCUGGCAGUUGCCACACAAUUGCUUGAGCACAUUAAGGUAUG---UCAUUU---U- .(((((((.(((.((......)).)))).((((((((.(..........).))))..((.((((((((.....)))))))).))........))))..)---))))).---.- ( -27.20) >DroGri_CAF1 55007 109 + 1 CAAUGGCACCGGCAUCAACUUAUCCCUCAAUUUGGGCGCGCCAAAUCCGGCGCCAAAUCUAGCUGUUGCCAAUCAGCUGCUGGAGCACAUUAAGGUACC---GUCUCCUACU- .((((((.(((((..........(((.......))).(((((......))))).......(((((........)))))))))).)).)))).(((....---....)))...- ( -34.70) >DroEre_CAF1 47580 109 + 1 CAACGGCACCGGCAUCAAUUUGUCCAUCAAUCUGGGCUCACCCAAUCCGGCGCCCAAUCUGGCGGUUGCCACACAGCUGCUCGAGCAUAUUAAGGUUAG---ACCUGCGAAU- ....(((.((((.........(((((......))))).........)))).)))...((.((((((((.....)))))))).))........(((....---.)))......- ( -32.87) >DroWil_CAF1 55819 108 + 1 CAAUGGUACUGGCAUCAAUUUGUCCCUCAAUCUGGGUGCUCCGAAUCCUGCGCCCAAUCUAGCAGUUGCCACACAAUUGCUCGAGCAUAUUAAGGUAAAGUGAUUUG-----G (((...((((((((......))))(((.(((.(((((((..........))))))).((.((((((((.....)))))))).))....))).)))...))))..)))-----. ( -29.90) >DroAna_CAF1 42660 106 + 1 CAACGGCACCGGCAUCAACUUGUCUAUCAACCUGGGCGCCCCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACUCAACUCCUUGAGCAUAUUAAGGUAAGG--AAUUA-----U ....((((((((((......))))........((((((((........)))))))).......)).))))..((....((((((.....))))))....)--)....-----. ( -36.50) >consensus CAACGGCACCGGCAUCAAUUUGUCCAUCAAUCUGGGCGCACCCAAUCCGGCGCCCAAUCUAGCGGUUGCCACACAACUGCUCGAGCAUAUUAAGGUAAG___ACUUG______ ....(((.((((.........((((........)))).........)))).)))...((.((((((((.....)))))))).))............................. (-22.39 = -22.17 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:43 2006