
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,236,072 – 5,236,220 |
| Length | 148 |
| Max. P | 0.966402 |

| Location | 5,236,072 – 5,236,182 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -12.58 |
| Energy contribution | -14.42 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
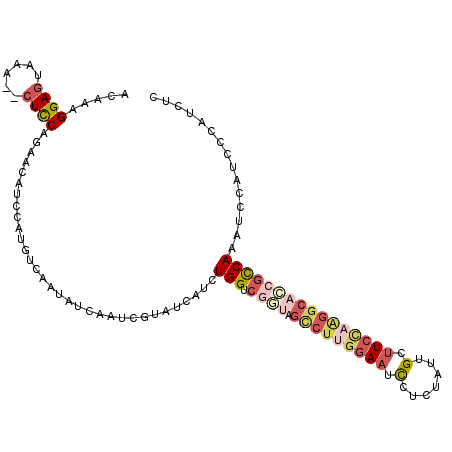
>3R_DroMel_CAF1 5236072 110 - 27905053 AUCGUAUCAUCUGGUCGGUAGCCUUGGAAUCCUCUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUCGAAUUUUAGCCGUAUAUUCGUUUAUCUAUGUAAGUACUA ...((((....(((.((((.(((((((((..(......).))))))))))))))))....(((.......(((((...........))))).......)))...)))).. ( -27.84) >DroSec_CAF1 9830 102 - 1 AUCGUAUCAUCUGGUCGGUAGCCUUGGAAUCCCCUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUCGAAUUUUAGCGGUAUAUUCGUUUAUCU--------ACUA ...(((.....(((.((((.(((((((((.(.......).))))))))))))))))......................(((((.....)))))....)--------)).. ( -26.70) >DroSim_CAF1 9888 110 - 1 AUCGUAUCAUCUGGUCGGUAGCCUUGGAAUUUACUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUCGAAUUUUAGCCGUGUAUUCUUUUAUCUAUGUAAGUACUA ...((((....(((.((((.(((((((((...........))))))))))))))))...............((((...........))))..............)))).. ( -25.50) >DroEre_CAF1 12107 106 - 1 AUCGUAUCAUCUGGUCAAAAGCCUUGGAAUCCUCUAUUGGUUCUUAGGCACCGCCAAAUCCAUCCCAUCUCCAAUUUUAGCUGUAUAUUCGAUUAUCU----AAGUACUA ...((((....(((((....((((.((((.((......)))))).))))((.((.(((.................))).)).))......)))))...----..)))).. ( -17.93) >DroYak_CAF1 9856 105 - 1 AUCGUAUCAUCUGGUCGUUAGUCUUGGAAUCCUCUAUUGGUUCUAGGGCAACGCCAAAUCCAUCCCAUCUCCAAUUUU-GCCGUAUAUUCGAUUAUCU----AAGUACUA ...((((....(((.((((.(((((((((.((......))))))))))))))))))......................-..((......)).......----..)))).. ( -22.20) >DroAna_CAF1 9430 97 - 1 AGUAUAUAACCUGGCUCA---------AAGCCUCUAUUGGCUCUCUGGCCGAUUCAUCCACAUCCCAUCUAGAUAUUUAGCUACGUAAUGUUUUAACU----AAGUAAUA (((((((.((.(((((.(---------(((((......)))).(((((..(((........)))....)))))..)).))))).)).))))....)))----........ ( -14.70) >consensus AUCGUAUCAUCUGGUCGGUAGCCUUGGAAUCCUCUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUCGAAUUUUAGCCGUAUAUUCGUUUAUCU____AAGUACUA ...........(((.((((.(((((((((.(.......).))))))))))))))))...................................................... (-12.58 = -14.42 + 1.84)



| Location | 5,236,111 – 5,236,220 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -16.83 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5236111 109 - 27905053 ACAAAGGAGUAAA--AUCCAGGACAUCCAUGUCAAUAUCAAUCGUAUCAUCUGGUCGGUAGCCUUGGAAUCCUCUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUC .....(((.....--......((((....))))..................(((.((((.(((((((((..(......).))))))))))))))))..))).......... ( -30.60) >DroSec_CAF1 9861 109 - 1 ACAAAGGAGUAAA--CUCCAGGACAUCCAUGUCAAUAUCAAUCGUAUCAUCUGGUCGGUAGCCUUGGAAUCCCCUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUC .....((((....--))))..((((....))))..................(((.((((.(((((((((.(.......).))))))))))))))))............... ( -32.90) >DroSim_CAF1 9927 109 - 1 ACAAAGGAGUAAA--CUCCAGGACAUCCAUGUCAAUAUUAAUCGUAUCAUCUGGUCGGUAGCCUUGGAAUUUACUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUC .....((((....--))))..((((....))))..................(((.((((.(((((((((...........))))))))))))))))............... ( -32.80) >DroEre_CAF1 12142 109 - 1 ACAAAGGAGGAAA--CUUCACAACAUCCAUGUCAAUAUCAAUCGUAUCAUCUGGUCAAAAGCCUUGGAAUCCUCUAUUGGUUCUUAGGCACCGCCAAAUCCAUCCCAUCUC .....((((....--))))................................((((.....((((.((((.((......)))))).))))...))))............... ( -21.70) >DroYak_CAF1 9890 109 - 1 ACAAAGGAGAAAA--CUCCACAGCAUCCAUGUCUAUAUCAAUCGUAUCAUCUGGUCGUUAGUCUUGGAAUCCUCUAUUGGUUCUAGGGCAACGCCAAAUCCAUCCCAUCUC .....((((....--))))................................(((.((((.(((((((((.((......))))))))))))))))))............... ( -25.80) >DroAna_CAF1 9465 101 - 1 ACAAAGGAGAACAAUCUCCA-AAGACCCAUACCAAUAUCAAGUAUAUAACCUGGCUCA---------AAGCCUCUAUUGGCUCUCUGGCCGAUUCAUCCACAUCCCAUCUA .....(((((....))))).-.(((...((((.........))))......(((((..---------.((((......))))....)))))................))). ( -19.10) >consensus ACAAAGGAGUAAA__CUCCAGAACAUCCAUGUCAAUAUCAAUCGUAUCAUCUGGUCGGUAGCCUUGGAAUCCUCUAUUGCUUCCAAGGCACCGCCAAAUCCAUCCCAUCUC .....((((......))))................................(((.((((.(((((((((.(.......).))))))))))))))))............... (-16.83 = -18.70 + 1.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:33 2006