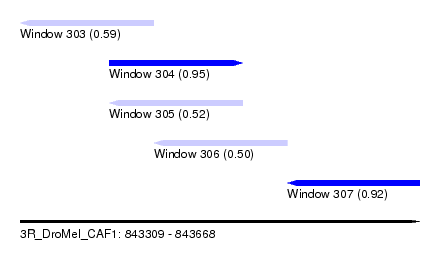
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 843,309 – 843,668 |
| Length | 359 |
| Max. P | 0.945983 |

| Location | 843,309 – 843,429 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -29.75 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 843309 120 - 27905053 AGACAUGACUGUAACUUUGUCGUGGCUGCCACUUCCACUUCCUUUUCCUCCUCCACUUGCGAUUCCCCUACGUCCUGGCAGCCAUGUCCUCAUUGAGCUGUUCGCCUAAGUGGGCGACUG .(((..(((.........)))((((((((((..............(((..........).)).............))))))))))))).............((((((....))))))... ( -29.63) >DroSec_CAF1 19083 120 - 1 AGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACCUCCUUUUCCCCCGCCACUGGCAAUUCCCCUACGUCCUGGCAGCCAUGUCCUCAUUGAGCUGUUCGCCUAAGUGGGCGACUG .((((((((((.....((((((((((.........................))))).)))))....((........)))))))))))).............((((((....))))))... ( -37.91) >DroSim_CAF1 18296 120 - 1 AGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACCUCCUUUUCCCCCGCCACUUGCAAUUCCCCUACGUCCUGGCAGCCAUGUCCUCAUUGAGCUGUUCGCCUAAGUGGGCGACUG .((((((((((.....((((.(((((.........................)))))..))))....((........)))))))))))).............((((((....))))))... ( -34.81) >DroEre_CAF1 20031 104 - 1 AGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACUUCCUCCUCCAC----------------CUUCCGUCUUGGCAGCCAUGUCCUCAUUGAGCUGUUCGCCUAAGUGGGCGACUG ....((((((.(((....(.(((((((((((...................----------------.........))))))))))).)....)))))))))((((((....))))))... ( -31.59) >consensus AGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACCUCCUUUUCCCCCGCCACUUGCAAUUCCCCUACGUCCUGGCAGCCAUGUCCUCAUUGAGCUGUUCGCCUAAGUGGGCGACUG .((((.((......)).))))((((((((((............................................))))))))))(((((((((..((.....))...)))))).))).. (-29.75 = -30.00 + 0.25)

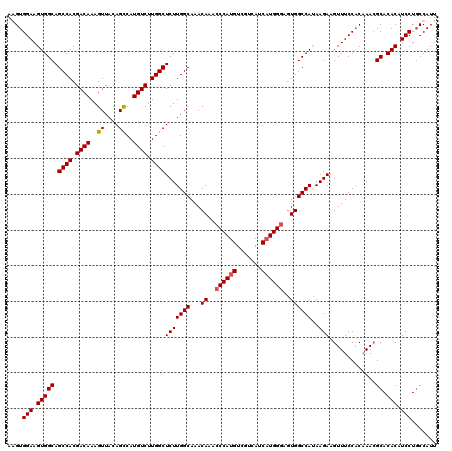
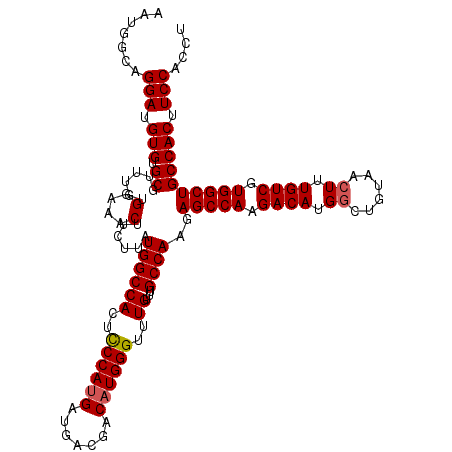
| Location | 843,389 – 843,509 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -33.86 |
| Energy contribution | -34.17 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 843389 120 + 27905053 AAGUGGAAGUGGCAGCCACGACAAAGUUACAGUCAUGUCUUGGCUCUUGGCAAACAAACCCAUGUCGUCAUCAUGGAAAUGGCCAUAAGAAGUUUCCACAAACGCACACAUCCUGCCAUU ..((((((((((((((((.((((............)))).)))))..............(((((.......))))).....))))).......))))))....(((.......))).... ( -31.01) >DroSec_CAF1 19163 120 + 1 AGGUGGAAGUGGCAGCCACGACAAAGUUACAGCCAUGUCUUGGCUCUUGGCAAACAAACCCAUGUCGUCAUCAUGGGAGUGGCCAUAAGAAGUUUCCACAAAGGCACACAUCCUGCCAUU ..((((((((((((((((.((((..((....))..)))).))))).........((..((((((.......))))))..))))))).......))))))...((((.......))))... ( -41.81) >DroSim_CAF1 18376 120 + 1 AGGUGGAAGUGGCAGCCACGACAAAGUUACAGCCAUGUCUUGGCUCUUGGCAAACAAACCCAUGUCGUCAUCAUGGGAGUGGCCAUAAGAAGUUUCCACAAACGCACACAUCCUGCCAUU .((((((.(((((.(((((......((((.(((((.....)))))..)))).......((((((.......)))))).)))))........((((....)))))).))).))).)))... ( -41.90) >DroEre_CAF1 20095 120 + 1 AAGUGGAAGUGGCAGCCACGACAAAGUUACAGCCAUGUCUUGGCUCUUGGCAAACAAGCCCAUGUCGUCAUCCUGGGAGUGGCCACAAGAAGUUUCCCCGAACGCACACUUCCUUCCAUU ....(((((((((.(((((((((.......(((((.....)))))...(((......)))..))))....((....)))))))........((((....)))))).)))))))....... ( -39.50) >consensus AAGUGGAAGUGGCAGCCACGACAAAGUUACAGCCAUGUCUUGGCUCUUGGCAAACAAACCCAUGUCGUCAUCAUGGGAGUGGCCAUAAGAAGUUUCCACAAACGCACACAUCCUGCCAUU ....(((.(((((.((((.((((..((....))..)))).))))(((((((...((..((((((.......))))))..))))))..))).............)).))).)))....... (-33.86 = -34.17 + 0.31)

| Location | 843,389 – 843,509 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -36.44 |
| Energy contribution | -36.75 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 843389 120 - 27905053 AAUGGCAGGAUGUGUGCGUUUGUGGAAACUUCUUAUGGCCAUUUCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCAAGACAUGACUGUAACUUUGUCGUGGCUGCCACUUCCACUU .......(((.(((.((......(....)......((((((..((((((.......))))))..))...))))..(((((.((((............)))).)))))))))).))).... ( -35.90) >DroSec_CAF1 19163 120 - 1 AAUGGCAGGAUGUGUGCCUUUGUGGAAACUUCUUAUGGCCACUCCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCAAGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACCU ...((((.......))))...((((((........((((((..((((((.......))))))..))...))))..(((((.((((.((......)).)))).))))).....)))))).. ( -39.60) >DroSim_CAF1 18376 120 - 1 AAUGGCAGGAUGUGUGCGUUUGUGGAAACUUCUUAUGGCCACUCCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCAAGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACCU ...((..(((.(((.((......(....)......((((((..((((((.......))))))..))...))))..(((((.((((.((......)).)))).)))))))))).))).)). ( -39.70) >DroEre_CAF1 20095 120 - 1 AAUGGAAGGAAGUGUGCGUUCGGGGAAACUUCUUGUGGCCACUCCCAGGAUGACGACAUGGGCUUGUUUGCCAAGAGCCAAGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACUU .......(((((((.((....(((....))).....((((((((....)).(((((....(((......))).(.(((((.....))))).)....)))))))))))))))))))).... ( -43.60) >consensus AAUGGCAGGAUGUGUGCGUUUGUGGAAACUUCUUAUGGCCACUCCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCAAGACAUGGCUGUAACUUUGUCGUGGCUGCCACUUCCACCU .......(((.(((.((......(....)......((((((..((((((.......))))))..))...))))..(((((.((((.((......)).)))).)))))))))).))).... (-36.44 = -36.75 + 0.31)

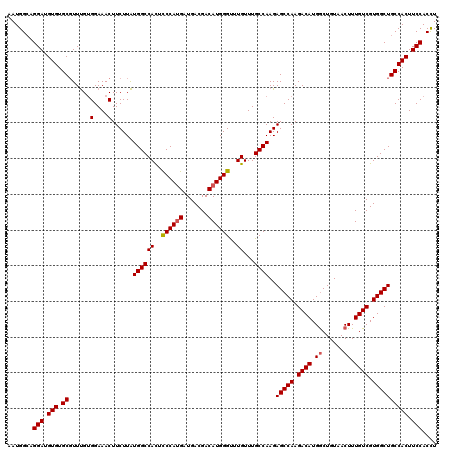
| Location | 843,429 – 843,549 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -40.69 |
| Consensus MFE | -34.22 |
| Energy contribution | -36.10 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 843429 120 - 27905053 GCUGCAUUUGCAGGGCUCACUCACACGACUGCACAUGGAAAAUGGCAGGAUGUGUGCGUUUGUGGAAACUUCUUAUGGCCAUUUCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCA .((((....))))(((((((.((((((.((((.(((.....)))))))..)))))).))....(....)......((((((..((((((.......))))))..))...)))).))))). ( -45.50) >DroSec_CAF1 19203 120 - 1 GCUGCAUUUGCUGGGCUCACUCACACGACUGCACAUGGAAAAUGGCAGGAUGUGUGCCUUUGUGGAAACUUCUUAUGGCCACUCCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCA ...((....))..(((((...((((((.((((.(((.....)))))))..)))))).......(....)......((((((..((((((.......))))))..))...)))).))))). ( -41.00) >DroSim_CAF1 18416 120 - 1 GCUGCAUUUCCUGGGCUCACUCACACGACUGCACAUGGAAAAUGGCAGGAUGUGUGCGUUUGUGGAAACUUCUUAUGGCCACUCCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCA .............(((((((.((((((.((((.(((.....)))))))..)))))).))....(....)......((((((..((((((.......))))))..))...)))).))))). ( -41.80) >DroEre_CAF1 20135 120 - 1 GCUGCAGUUGCAGGACUCGCUCAAAGGACUGCACAUGGAAAAUGGAAGGAAGUGUGCGUUCGGGGAAACUUCUUGUGGCCACUCCCAGGAUGACGACAUGGGCUUGUUUGCCAAGAGCCA .((((....)))).....((((...((((.((((((...............)))))))))).((.((((..((..((..((.((....))))....))..))...)))).))..)))).. ( -34.46) >consensus GCUGCAUUUGCAGGGCUCACUCACACGACUGCACAUGGAAAAUGGCAGGAUGUGUGCGUUUGUGGAAACUUCUUAUGGCCACUCCCAUGAUGACGACAUGGGUUUGUUUGCCAAGAGCCA .((((....))))(((((((.(((((..((((.(((.....)))))))...))))).))....(....)......((((((..((((((.......))))))..))...)))).))))). (-34.22 = -36.10 + 1.88)

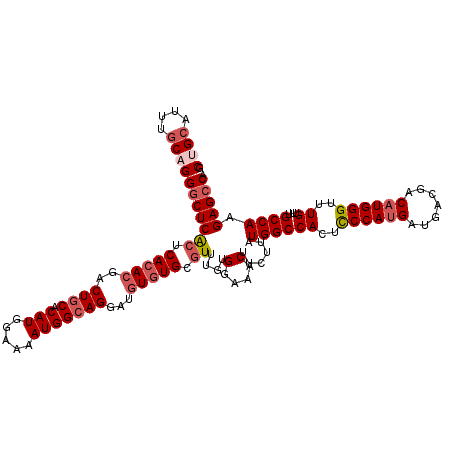
| Location | 843,549 – 843,668 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.92 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -23.29 |
| Energy contribution | -25.35 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 843549 119 - 27905053 AUGGUUAAUUUAGGUUUGCAUAAAUUAAGUUUCCCAUAUGGCUAUCUGGAAAUAUCCUGUCCAGCCAAGACACCA-AUGUCCAGGCUGCGGAGCUUUUCCUCCCCGGUAUGUUGGUAGCA ((((..(((((((.((.....)).)))))))..))))...((((((.....(((((.....(((((..((((...-.))))..))))).((((......))))..)))))...)))))). ( -33.60) >DroSec_CAF1 19323 98 - 1 AAGAUAAAUUUAGG----------------------UAUGGCUAUCUGGAAAUAUCCUGUCCAGCCAAGACACCAAAUGUCCAGGCUGCGGAGCUUUUCCUCCUCGGGAUGUUGGUAGCA ..............----------------------....((((((....(((((((((..(((((..((((.....))))..))))).((((......)))).))))))))))))))). ( -33.20) >DroSim_CAF1 18536 116 - 1 AAGGUAAAUUUAGG----CAUAAAUUAAGUUGCCCAUAUGGCUAUCUGGAAAUAUCCUGUCCAGCCAAGACACCAAAUGUCCAGGCUGCGGAGCUUUUCCUCCUCGGGAUGUGGGUAGCA ..((..(((((((.----......)))))))..)).....(((((((....((((((((..(((((..((((.....))))..))))).((((......)))).))))))))))))))). ( -38.90) >DroEre_CAF1 20255 107 - 1 AUAGCAAACUCAAGUUCGCAUAAAUUAAGUU------------GUCUGGAAAUAUCCUGACCAGCCAAGACACCAAGUAUCCA-GCUGCGGAACUUUUCCUCCCCGGGAUGUUAGUAGCA ...((.(((....))).)).........(((------------(((.(((....))).)).))))..................-(((((..(((...((((....)))).))).))))). ( -23.50) >consensus AAGGUAAAUUUAGG____CAUAAAUUAAGUU_____UAUGGCUAUCUGGAAAUAUCCUGUCCAGCCAAGACACCAAAUGUCCAGGCUGCGGAGCUUUUCCUCCCCGGGAUGUUGGUAGCA ........................................((((((....(((((((((..(((((..((((.....))))..))))).((((......)))).))))))))))))))). (-23.29 = -25.35 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:31 2006