
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,222,491 – 5,222,611 |
| Length | 120 |
| Max. P | 0.837600 |

| Location | 5,222,491 – 5,222,590 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.49 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -12.28 |
| Energy contribution | -11.73 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
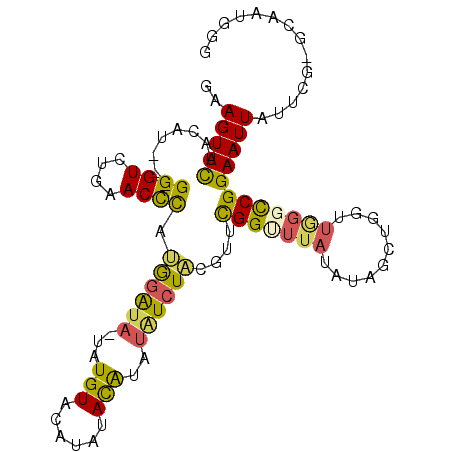
>3R_DroMel_CAF1 5222491 99 + 27905053 GAAGUUCAACAU---GGGUCUGAACUCAUGGAUA-UAUGUACAUGCACAUAUAUCUAUGUUCGGUUUAUAUAGCUGGUUGGCCCGGAAUUAUUCG-GCAAUGGG (((........(---(((((......((((((((-(((((......))))))))))))).((((((.....))))))..))))))......))).-........ ( -28.64) >DroSec_CAF1 5878 97 + 1 GAAGUUCAACAU---GGGUCUGAACUCAUGGAUA-UAUGUACAA--ACAUAUAUCUACGUUCGGUUUAUAUAGCUGGUUGGGCCGGAAUUAUUCG-GCAAUGGA .....(((((((---((((....))))))(((((-(((((....--)))))))))).....(((((.....))))))))))((((((....))))-))...... ( -29.50) >DroSim_CAF1 11213 97 + 1 GAAGUUCAACAU---GGGUCUGAACCCAUGGAUA-UAUGUACAU--ACAUAUAUCUACGUUCGGUUUAUAUAGCUGGUUGGGCCGGAAUUAUUCG-GCAAUGGA .....(((((((---((((....))))))(((((-(((((....--)))))))))).....(((((.....))))))))))((((((....))))-))...... ( -32.50) >DroEre_CAF1 11991 95 + 1 GAAGUUCAACAU---GGGUCUGAACUCACAGAUA-UAUGUAGCUAUAUA----UCUGCGUUCGGCUUAUAUAGCUGGCUAGGCCGGAAUUAUUCG-GCAGUGGG ........((((---(.(((((......))))).-)))))(((((((((----...((.....)).)))))))))..(((.((((((....))))-))..))). ( -30.00) >DroYak_CAF1 6271 89 + 1 GAAGUUUAGCAU---GGGUGUGAACCCAUAGAUA-CAUGUAGUCAUAUAUCUAUCUACGUUCGGUUUAUAUAGCUCGCUGGGCCGGAAUUAUU----------- ...(((((((..---(.((((((((((.((((((-.(((((....))))).)))))).)...)))))))))..)..)))))))..........----------- ( -23.60) >DroAna_CAF1 6011 85 + 1 GAAGUUCAAGAAGUGGGGUCGGAACCUCAGAGUAAAUUGUAUAUAUACGGAUGUCU-----UGG--------------UCUUUCGGAAUUGUUCGGGUAAUGGG ((((..(((((..((((((....)))))).......(((((....)))))...)))-----)).--------------.))))((((....))))......... ( -16.10) >consensus GAAGUUCAACAU___GGGUCUGAACCCAUGGAUA_UAUGUACAUAUACAUAUAUCUACGUUCGGUUUAUAUAGCUGGUUGGGCCGGAAUUAUUCG_GCAAUGGG ..(((((........((((....)))).((((((...(((......)))..))))))....(((((((..........)))))))))))).............. (-12.28 = -11.73 + -0.55)


| Location | 5,222,518 – 5,222,611 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -24.01 |
| Consensus MFE | -18.77 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5222518 93 + 27905053 GAUAUAUGUACAUGCACAUAUAUCUAUGUUCGGUUUAUAUAGCUGGUUGGCCCGGAAUUAUUCGGCAAUGGGAACUUGAUGAACAA---UAGAUGA (((((((((......)))))))))..((((((..........((.((((..(((((....))))))))).)).......)))))).---....... ( -22.53) >DroSec_CAF1 5905 91 + 1 GAUAUAUGUACAA--ACAUAUAUCUACGUUCGGUUUAUAUAGCUGGUUGGGCCGGAAUUAUUCGGCAAUGGAAACUCGAGGAACAA---UAGAUGA (((((((((....--)))))))))..(((((((((.....)))))((((.((((((....))))))...(....)........)))---).)))). ( -24.40) >DroSim_CAF1 11240 91 + 1 GAUAUAUGUACAU--ACAUAUAUCUACGUUCGGUUUAUAUAGCUGGUUGGGCCGGAAUUAUUCGGCAAUGGAAACUCGAGGAACAA---UAGAUGA (((((((((....--)))))))))..(((((((((.....)))))((((.((((((....))))))...(....)........)))---).)))). ( -24.40) >DroEre_CAF1 12018 92 + 1 GAUAUAUGUAGCUAUAUA----UCUGCGUUCGGCUUAUAUAGCUGGCUAGGCCGGAAUUAUUCGGCAGUGGGAACUCGACGAACAAUAAUAGUUGA ........((((((((((----...((.....)).)))))))))).(((.((((((....))))))..)))....(((((...........))))) ( -24.70) >consensus GAUAUAUGUACAU__ACAUAUAUCUACGUUCGGUUUAUAUAGCUGGUUGGGCCGGAAUUAUUCGGCAAUGGAAACUCGAGGAACAA___UAGAUGA (((((((((......)))))))))...((((((((.....))))......((((((....))))))..............))))............ (-18.77 = -19.65 + 0.88)



| Location | 5,222,518 – 5,222,611 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -17.94 |
| Consensus MFE | -11.85 |
| Energy contribution | -13.59 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5222518 93 - 27905053 UCAUCUA---UUGUUCAUCAAGUUCCCAUUGCCGAAUAAUUCCGGGCCAACCAGCUAUAUAAACCGAACAUAGAUAUAUGUGCAUGUACAUAUAUC .......---.(((((...............(((........)))((......))..........)))))..((((((((((....)))))))))) ( -18.30) >DroSec_CAF1 5905 91 - 1 UCAUCUA---UUGUUCCUCGAGUUUCCAUUGCCGAAUAAUUCCGGCCCAACCAGCUAUAUAAACCGAACGUAGAUAUAUGU--UUGUACAUAUAUC ...((((---(.((((....((((......((((........))))......)))).........)))))))))(((((((--....))))))).. ( -17.32) >DroSim_CAF1 11240 91 - 1 UCAUCUA---UUGUUCCUCGAGUUUCCAUUGCCGAAUAAUUCCGGCCCAACCAGCUAUAUAAACCGAACGUAGAUAUAUGU--AUGUACAUAUAUC ...((((---(.((((....((((......((((........))))......)))).........)))))))))(((((((--....))))))).. ( -17.02) >DroEre_CAF1 12018 92 - 1 UCAACUAUUAUUGUUCGUCGAGUUCCCACUGCCGAAUAAUUCCGGCCUAGCCAGCUAUAUAAGCCGAACGCAGA----UAUAUAGCUACAUAUAUC .........((((((((.(.(((....)))).))))))))...(((...)))(((((((((.((.....))...----)))))))))......... ( -19.10) >consensus UCAUCUA___UUGUUCCUCGAGUUCCCAUUGCCGAAUAAUUCCGGCCCAACCAGCUAUAUAAACCGAACGUAGAUAUAUGU__AUGUACAUAUAUC ...........(((((....((((......((((........))))......)))).........)))))..((((((((((....)))))))))) (-11.85 = -13.59 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:25 2006