
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,212,569 – 5,212,661 |
| Length | 92 |
| Max. P | 0.803678 |

| Location | 5,212,569 – 5,212,661 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
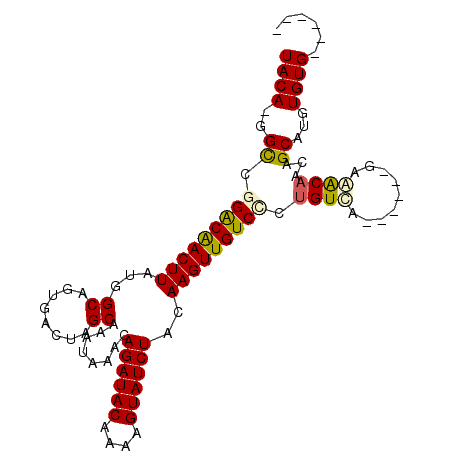
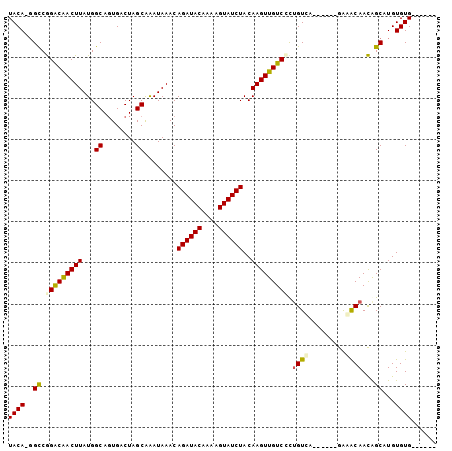
>3R_DroMel_CAF1 5212569 92 + 27905053 UACA-GGCCGGACAACUUAUGGCAGUGACUAGCGAAUAAACAGAUACAAAAGUAUCUACAAGUUGUCCCUGUCAA-----GAGACAACGGCAUGUGUG------ ((((-.((((((((((((...((........))........((((((....))))))..))))))))).((((..-----..))))..))).))))..------ ( -31.00) >DroPse_CAF1 19774 100 + 1 UACAUGGCCGGGCAACUUAUGGCAGUGACUAGCAGAUAAACAGAUACAAAAGUAUCUACAAGUUGUCUUUGUGU-UCGCUGGAGCAACAGCAUGUGUGUGU--- ((((((...(((((((((...((........))........((((((....))))))..)))))))))((((((-((....)))).)))))))))).....--- ( -27.00) >DroSec_CAF1 18433 92 + 1 UACA-GGCCGGACAACUUAUGGCACUGACUAGCGAAUAAACAGAUACAAAAGUAUCUACAAGUUGUCCCUGUCAA-----GAGACAACGGCAUGUGUG------ ((((-.((((((((((((...((........))........((((((....))))))..))))))))).((((..-----..))))..))).))))..------ ( -31.00) >DroYak_CAF1 19633 92 + 1 UACA-GGCCGGACAACUUAUGGCAGUGACUAGCGAAUAAACAGAUACAAAAGUAUCUACAAGUUGUCGCUGUCAG-----GAGACAACAGCAUGUGUG------ ((((-.((.(.....(((.((((((((((.(((........((((((....))))))....))))))))))))).-----)))....).)).))))..------ ( -28.10) >DroAna_CAF1 18926 103 + 1 UACA-UGUCAGGCGACUUAUGGCAGUGACUAGCAGAUAAACAGAUACCAAAGUAUCUACAAGUUGUCCCCGUUCGCCGCUAAAGCAAUAGCAUGUGUGUGCGCC ((((-(((..(((((...((((..(..(((.(.(((((....(....)....))))).).)))..)..)))))))))((....))....)))))))........ ( -27.30) >DroPer_CAF1 19618 99 + 1 UACAUGGCCGGGCAACUUAUGGCAGUGACUAGCAGAUAAACAGAUACAAAAGUAUCUACAAGUUGUCUUUGUGU-UCGCUGGAGCAACAGCAUGUGUGUG---- ((((((...(((((((((...((........))........((((((....))))))..)))))))))((((((-((....)))).))))))))))....---- ( -27.00) >consensus UACA_GGCCGGACAACUUAUGGCAGUGACUAGCAAAUAAACAGAUACAAAAGUAUCUACAAGUUGUCCCUGUCA______GAAACAACAGCAUGUGUG______ ((((..((.(((((((((...((........))........((((((....))))))..))))))))).((((.........))))...))...))))...... (-19.90 = -19.68 + -0.22)

| Location | 5,212,569 – 5,212,661 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -17.09 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
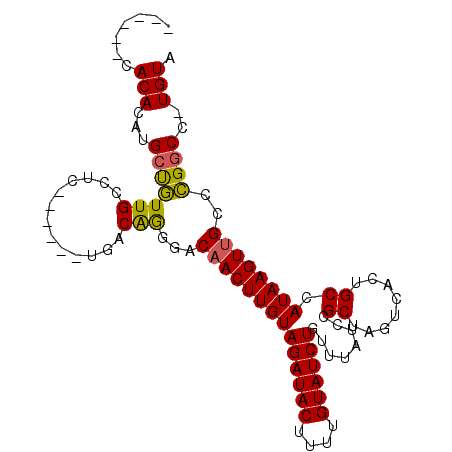
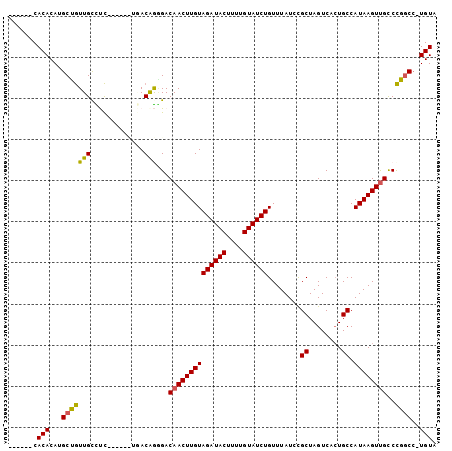
>3R_DroMel_CAF1 5212569 92 - 27905053 ------CACACAUGCCGUUGUCUC-----UUGACAGGGACAACUUGUAGAUACUUUUGUAUCUGUUUAUUCGCUAGUCACUGCCAUAAGUUGUCCGGCC-UGUA ------...(((.(((.(((((..-----..)))))(((((((((((((((((....))))))........((........)).)))))))))))))).-))). ( -34.90) >DroPse_CAF1 19774 100 - 1 ---ACACACACAUGCUGUUGCUCCAGCGA-ACACAAAGACAACUUGUAGAUACUUUUGUAUCUGUUUAUCUGCUAGUCACUGCCAUAAGUUGCCCGGCCAUGUA ---......((((((((((((....))))-.........((((((((((((((....))))))........((........)).))))))))..))).))))). ( -24.00) >DroSec_CAF1 18433 92 - 1 ------CACACAUGCCGUUGUCUC-----UUGACAGGGACAACUUGUAGAUACUUUUGUAUCUGUUUAUUCGCUAGUCAGUGCCAUAAGUUGUCCGGCC-UGUA ------...(((.(((.(((((..-----..)))))(((((((((((((((((....)))))).......((((....))))..)))))))))))))).-))). ( -35.20) >DroYak_CAF1 19633 92 - 1 ------CACACAUGCUGUUGUCUC-----CUGACAGCGACAACUUGUAGAUACUUUUGUAUCUGUUUAUUCGCUAGUCACUGCCAUAAGUUGUCCGGCC-UGUA ------...(((.(((((((((..-----..))))))((((((((((((((((....))))))........((........)).)))))))))).))).-))). ( -32.00) >DroAna_CAF1 18926 103 - 1 GGCGCACACACAUGCUAUUGCUUUAGCGGCGAACGGGGACAACUUGUAGAUACUUUGGUAUCUGUUUAUCUGCUAGUCACUGCCAUAAGUCGCCUGACA-UGUA ((((.((......((((......))))((((.(((((.....)))))((((((....)))))).................))))....)))))).....-.... ( -27.70) >DroPer_CAF1 19618 99 - 1 ----CACACACAUGCUGUUGCUCCAGCGA-ACACAAAGACAACUUGUAGAUACUUUUGUAUCUGUUUAUCUGCUAGUCACUGCCAUAAGUUGCCCGGCCAUGUA ----.....((((((((((((....))))-.........((((((((((((((....))))))........((........)).))))))))..))).))))). ( -24.00) >consensus ______CACACAUGCUGUUGCCUC______UGACAGGGACAACUUGUAGAUACUUUUGUAUCUGUUUAUCCGCUAGUCACUGCCAUAAGUUGCCCGGCC_UGUA .......(((...(((((((.............)))...((((((((((((((....))))))........((........)).))))))))..))))..))). (-17.09 = -16.57 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:21 2006