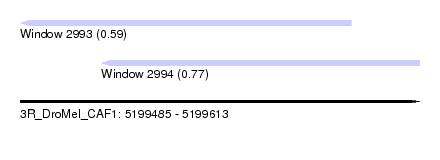
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,199,485 – 5,199,613 |
| Length | 128 |
| Max. P | 0.774636 |

| Location | 5,199,485 – 5,199,591 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -22.05 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
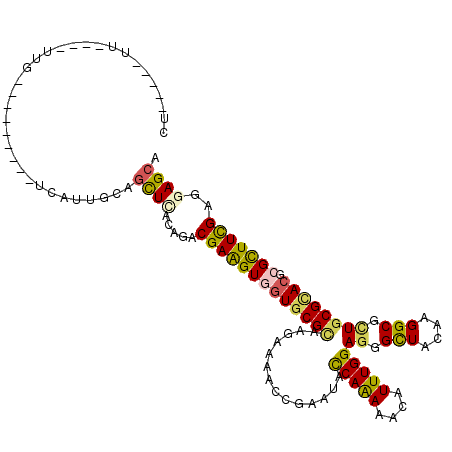
>3R_DroMel_CAF1 5199485 106 - 27905053 GACGAGGUGGUGCGCAAGAAAUCUGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGCUGCGCACGCGCUUCGAGGAGAACUACAUAAAGGGUGA-----------GUGAGA---AAUGA ..(((((((((((((..............(((((....)))))((.(((....))).)))))))).)))))))......(((.(((.....))))-----------))....---..... ( -33.30) >DroPse_CAF1 5720 117 - 1 GACGAGGUGCUGCGCAAGAAGACCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGCUGCGCACGCGUUUCGAGGAGCACUACAUUAAAGGUAACCCGAUCCCGACUGAGA---AAUGG ..(((..((((((((..............(((((....)))))((.(((....))).))))))).)))..))).(((....(((.......)))......))).........---..... ( -29.20) >DroGri_CAF1 7217 108 - 1 GACGAAGUUGUGCGUAAGAAGGGCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGUUGCGCACGCGCUUCGAAGAGCACUACAUAAAAGGUGA-----------GUGCCCC-CAUUGG ..(((((((((((((((............(((((....)))))...(((....))).))))))))).))))))....(((((.(((.....))))-----------))))...-...... ( -33.90) >DroWil_CAF1 6103 106 - 1 GACGAAGUGGUGCGUAAGAAGAGCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGUUGCGUACACGUUUCGAGGAGCACUAUAUAAAGGGUAA-----------GCCUCC---AUUCC ..(((((((((((((((............(((((....)))))...(((....))).)))))))).))))))).((((((((........)))..-----------).))))---..... ( -26.80) >DroMoj_CAF1 8163 109 - 1 GACGAAGUUGUGCGCAAGAAGAGCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGUUACGCACGCGCUUUGAAGAGCACUACAUAAAAGGUGA-----------GUAACACCCAAUGA ......((.((((.(......(((...(.(((((....))))))..))).((((((((.......))))))))..).)))).))......((((.-----------....))))...... ( -26.70) >DroAna_CAF1 5520 106 - 1 GACGAGGUGGUUCGCAAGAAAUCCGAAUAUCAGAAACAUUUGGAAGGCUACAAGGCUCUGCGCACGCGCUUCGAGGAGCACUAUAUUAAAGGUAA-----------UUUAAU---AUUUA ..(((((((((.((((.....(((((((.........))))))).((((....)))).)))).)).))))))).........((((((((.....-----------))))))---))... ( -27.40) >consensus GACGAAGUGGUGCGCAAGAAGACCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGCUGCGCACGCGCUUCGAGGAGCACUACAUAAAAGGUAA___________GUGACA___AAUGA ..((((((((((((((.............(((((....)))))...(((....)))..))))))).)))))))............................................... (-22.05 = -21.72 + -0.33)


| Location | 5,199,511 – 5,199,613 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.48 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5199511 102 - 27905053 CC-----UC----UUG---------UCAUUCCAGCUCACCGACGAGGUGGUGCGCAAGAAAUCUGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGCUGCGCACGCGCUUCGAGGAGAA ((-----((----..(---------((.............)))((((((((((((..............(((((....)))))((.(((....))).)))))))).)))))))))).... ( -36.22) >DroVir_CAF1 6085 105 - 1 CU-----UU----UUG------UAUGCAUUGCAGCUUACAGACGAAGUUGUGCGAAAGAAAAGCGAAUACCAAAAACAUUUGGAGGGUUACAAGGCGCUGCGCACGCGCUUUGAAGAGCA ..-----.(----(((------((.((......)).))))))((((((((((((.......((((....(((((....))))).(.....)....)))).)))))).))))))....... ( -27.10) >DroGri_CAF1 7245 105 - 1 CU-----UU----UCU------UAUGCAUUGCAGUUGACAGACGAAGUUGUGCGUAAGAAGGGCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGUUGCGCACGCGCUUCGAAGAGCA ((-----((----(((------(((((((.((.(((....)))...)).))))))))((((.(((....(((((....))))).(.((.((.....)).)).).))).)))))))))).. ( -34.10) >DroWil_CAF1 6129 106 - 1 CU-----UUUUUUUUG---------CUUUUGCAGUUGACUGACGAAGUGGUGCGUAAGAAGAGCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGUUGCGUACACGUUUCGAGGAGCA ..-----.......((---------(((((.(((....))).(((((((((((((((............(((((....)))))...(((....))).)))))))).)))))))))))))) ( -29.00) >DroAna_CAF1 5546 112 - 1 CUUCUCUUU----UUGUAUUU----UUUUUCCAGCUCACCGACGAGGUGGUUCGCAAGAAAUCCGAAUAUCAGAAACAUUUGGAAGGCUACAAGGCUCUGCGCACGCGCUUCGAGGAGCA .........----........----........((((.....(((((((((.((((.....(((((((.........))))))).((((....)))).)))).)).)))))))..)))). ( -30.20) >DroPer_CAF1 5726 111 - 1 CU-----UU----GUGAUCUCUCACUCAUUUCAGCUCACUGACGAGGUGCUGCGCAAGAAGACCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGCUGCGCACGCGUUUCGAGGAGCA ..-----..----((((....))))........((((.((..(((..((((((((..............(((((....)))))((.(((....))).))))))).)))..))))))))). ( -36.40) >consensus CU_____UU____UUG_________UCAUUGCAGCUCACAGACGAAGUGGUGCGCAAGAAAACCGAAUACCAAAAACAUUUGGAGGGCUACAAGGCGCUGCGCACGCGCUUCGAGGAGCA .................................((((.....(((((((((((((..............(((((....)))))((.(((....))).)))))))).)))))))..)))). (-23.76 = -23.48 + -0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:17 2006