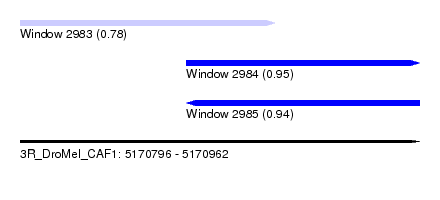
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,170,796 – 5,170,962 |
| Length | 166 |
| Max. P | 0.949033 |

| Location | 5,170,796 – 5,170,902 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5170796 106 + 27905053 AAUUUAAUGACCGCAGCUGUUUCUUUUGGGGACAGACUGUACUGCUUCGAUUUCGAUCUUGAUUAAAUCCCAUUGAAAAUAGGCAUUUAGAAUGCACAUAUAUCGG ..(((((((...((((((((..(.....)..)))).))))......(((....)))..............))))))).....((((.....))))........... ( -22.20) >DroSec_CAF1 9332 105 + 1 AAUGCAAUGACCGCAGCUGUUUCUUUUGGGGACAGACUGUACUGCUUCGAU-UCGAUCUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGG ...(((......((((((((..(.....)..)))).))))..)))..((((-.........((((((((((((.....))))).)))))))..........)))). ( -26.71) >DroSim_CAF1 9337 106 + 1 AAUGCAAUGACCGCAGCUGUUUCUUUUGGGGACAGACUGUACUGCUUCGAUUUCGAUCUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGG ..((((......((((((((..(.....)..)))).))))......(((....))).....((((((((((((.....))))).))))))).)))).......... ( -27.90) >DroEre_CAF1 9136 106 + 1 AAUUUAAUGACCGCAGCUGUUUGUUUUGGGGACAGACUGUACUGCUUCGAUUUCGAUCUUGAUUAAAUCCCAUUGUAAAUGGGCAUUUAGUAUGCACAUAUAUCGG ............((((..(((((((.....)))))))....))))..((((..........((((((((((((.....))))).)))))))..........)))). ( -25.25) >DroYak_CAF1 9547 105 + 1 AAAUUCAUGACCGCAGCUGUUUCUUUUGGGAACAGACUGUAC-GCUUCGAGUUCGAUCUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGG ..........(((((((((((((.....))))))).))))..-((..((((......))))((((((((((((.....))))).)))))))..)).........)) ( -28.30) >consensus AAUUUAAUGACCGCAGCUGUUUCUUUUGGGGACAGACUGUACUGCUUCGAUUUCGAUCUUGAUUAAAUCCCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGG ..........(((((((((((((.....))))))).))))...((.(((....))).....((((((((((((.....))))).)))))))..)).........)) (-24.50 = -24.94 + 0.44)



| Location | 5,170,865 – 5,170,962 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -21.20 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
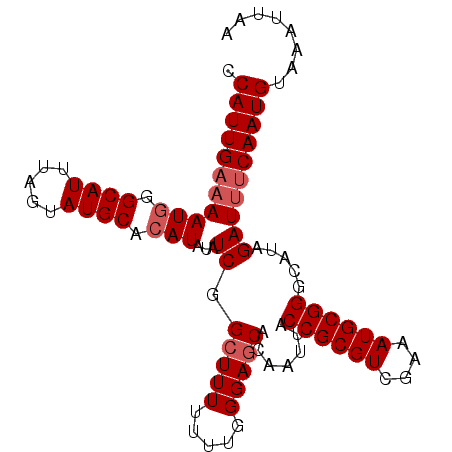
>3R_DroMel_CAF1 5170865 97 + 27905053 CCAUUGAAAAUAGGCAUUUAGAAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCCAAAUGCGGGCAUAGAUUUCAAUGUAAAUUAA .((((((((....((((.....))))...(((..(.((....(((((..((.........))..))))).)).)..)))..))))))))........ ( -23.10) >DroSec_CAF1 9400 97 + 1 CCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCGAAAUGCGGGCAUAGAUUUCAAUGUAAAUUAA .(((((((((((.((((.....)))).)))...((.(((((.....))))).......((((((....)))))).....))))))))))........ ( -24.80) >DroSim_CAF1 9406 97 + 1 CCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCGAAAUGCGGGCAUAGAUUUCAAUGUAAAUUAA .(((((((((((.((((.....)))).)))...((.(((((.....))))).......((((((....)))))).....))))))))))........ ( -24.80) >DroEre_CAF1 9205 96 + 1 CCAUUGUAAAUGGGCAUUUAGUAUGCACAUAUAUCGGGUUU-UUUGGGAGCACAAUUACCGCGUUGAAAUGCGGGCAUAGAUAUCAAUGUAAAUUAA .(((((...(((.((((.....)))).)))(((((..((((-.....)))).......((((((....)))))).....))))))))))........ ( -20.80) >DroYak_CAF1 9615 96 + 1 CCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUU-UUUGGGAGCACAAUUACCGCGUUGAAAUGCGGGCAUAGAUUUCAAUGUAAAUUAA .(((((((((((.((((.....)))).)))...((.(((((-....))))).......((((((....)))))).....))))))))))........ ( -24.90) >consensus CCAUUGAAAAUGGGCAUUUAGUAUGCACAUAUAUCGGCUUUUUUUGGGAGCACAAUUACCGCGUCGAAAUGCGGGCAUAGAUUUCAAUGUAAAUUAA .(((((((((((.((((.....)))).)))...((.(((((.....))))).......((((((....)))))).....))))))))))........ (-21.20 = -22.00 + 0.80)

| Location | 5,170,865 – 5,170,962 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
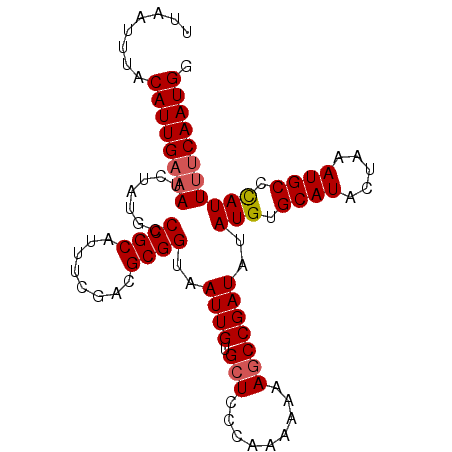
>3R_DroMel_CAF1 5170865 97 - 27905053 UUAAUUUACAUUGAAAUCUAUGCCCGCAUUUGGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUUCUAAAUGCCUAUUUUCAAUGG ........((((((((.........(((((((((((((...((((.(((.........)))))))...))))...)))))))))....)))))))). ( -22.72) >DroSec_CAF1 9400 97 - 1 UUAAUUUACAUUGAAAUCUAUGCCCGCAUUUCGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGG ........((((((((.......((((........))))..((((.(((.........)))))))..(((.((((.....)))).))))))))))). ( -20.50) >DroSim_CAF1 9406 97 - 1 UUAAUUUACAUUGAAAUCUAUGCCCGCAUUUCGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGG ........((((((((.......((((........))))..((((.(((.........)))))))..(((.((((.....)))).))))))))))). ( -20.50) >DroEre_CAF1 9205 96 - 1 UUAAUUUACAUUGAUAUCUAUGCCCGCAUUUCAACGCGGUAAUUGUGCUCCCAAA-AAACCCGAUAUAUGUGCAUACUAAAUGCCCAUUUACAAUGG ........((((((((((.....((((........))))...(((......))).-......)))))(((.((((.....)))).)))...))))). ( -17.50) >DroYak_CAF1 9615 96 - 1 UUAAUUUACAUUGAAAUCUAUGCCCGCAUUUCAACGCGGUAAUUGUGCUCCCAAA-AAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGG ........((((((((.......((((........))))..((((.(((......-..)))))))..(((.((((.....)))).))))))))))). ( -21.30) >consensus UUAAUUUACAUUGAAAUCUAUGCCCGCAUUUCGACGCGGUAAUUGUGCUCCCAAAAAAAGCCGAUAUAUGUGCAUACUAAAUGCCCAUUUUCAAUGG ........((((((((.......((((........))))..((((.(((.........)))))))..(((.((((.....)))).))))))))))). (-17.74 = -18.18 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:09 2006