
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,164,823 – 5,164,922 |
| Length | 99 |
| Max. P | 0.950675 |

| Location | 5,164,823 – 5,164,922 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -21.69 |
| Consensus MFE | -12.52 |
| Energy contribution | -14.13 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5164823 99 + 27905053 CUUCUCC-UUUUCGACUGCACAAUGCACUUCUUUUUCUUCUUCCGCAACCUUGUG-CGUGUGCCCACAUGUUUACAUAAAGUGCAACUGUGUGCACGAAAA .......-.(((((..((((((.(((((((..............(((......))-)(((((..(....)..))))).)))))))....))))))))))). ( -25.40) >DroPse_CAF1 3351 84 + 1 CUUUUCCCCAUAUGGCUGUACAAUGCACUCGCCC----UCACACUCAACCUUGUAGCAAACUUACAUAUAUGUACAUAGA---------UAUGU----AAA ........(((((...((((((..((....))..----.............(((((.....)))))....))))))...)---------)))).----... ( -10.20) >DroSec_CAF1 3445 99 + 1 CUUCUCC-UUUUCGCCUGCACAAUGCACUUCUUCUUCUUCAUCCACAACCUUGUG-AGUGUGCCCACAUGUUUACAUAAAGUGCAACUGUGUGCACGAAAA .......-.(((((..((((((.(((((((.............((((....))))-.(((((..(....)..))))).)))))))....))))))))))). ( -25.50) >DroSim_CAF1 3440 99 + 1 CUUCUCC-UUUUCGCCUGCACAAUGCACUUCUUCUUCUUCAUCCACAACCUUGUG-CGUGUGCCCACAUGUUUACAUAAAGUGCAACUGUGUGCACGAAAA .......-.(((((..((((((.(((((((.............((((....))))-(((((....)))))........)))))))....))))))))))). ( -25.50) >DroEre_CAF1 3098 99 + 1 CUUCUCC-UUUUCGCCUGUAAAAUGCACUUCUUCUUCUUCUGCCACAACCUUGUG-CAUGUGCCCACAUGUUUACAUAAAGUGCAACUGUGUGCACGAAAA .......-.((((.........((((((........................)))-)))((((.((((.((((((.....))).))))))).)))))))). ( -21.36) >DroYak_CAF1 3488 99 + 1 CUUCUCU-CUUUCGCCUGUACACUGCACUUCUUCUUCUUCUGCCACAACCUUGUG-CAUGUGCCCACAUGUUUACAUAAAGUGCAACUGUAUGCACGAAAA .......-.(((((..((((((.(((((((.............((((....))))-(((((....)))))........)))))))..))))))..))))). ( -22.20) >consensus CUUCUCC_UUUUCGCCUGCACAAUGCACUUCUUCUUCUUCAUCCACAACCUUGUG_CAUGUGCCCACAUGUUUACAUAAAGUGCAACUGUGUGCACGAAAA .........(((((..((((((.(((((((.............((((....)))).(((((....)))))........)))))))..))))))..))))). (-12.52 = -14.13 + 1.61)

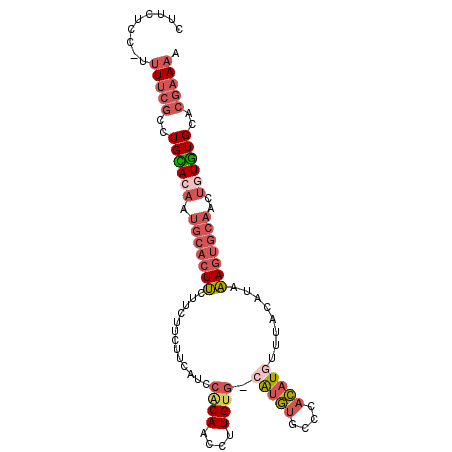

| Location | 5,164,823 – 5,164,922 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -13.65 |
| Energy contribution | -15.65 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5164823 99 - 27905053 UUUUCGUGCACACAGUUGCACUUUAUGUAAACAUGUGGGCACACG-CACAAGGUUGCGGAAGAAGAAAAAGAAGUGCAUUGUGCAGUCGAAAA-GGAGAAG ((((((((((((....((((((((.....(((.((((.(....).-))))..))).(....)........)))))))).))))))..))))))-....... ( -28.10) >DroPse_CAF1 3351 84 - 1 UUU----ACAUA---------UCUAUGUACAUAUAUGUAAGUUUGCUACAAGGUUGAGUGUGA----GGGCGAGUGCAUUGUACAGCCAUAUGGGGAAAAG ((.----.((((---------(...((((((...(((((.((((..((((........)))).----))))...)))))))))))...)))))..)).... ( -18.40) >DroSec_CAF1 3445 99 - 1 UUUUCGUGCACACAGUUGCACUUUAUGUAAACAUGUGGGCACACU-CACAAGGUUGUGGAUGAAGAAGAAGAAGUGCAUUGUGCAGGCGAAAA-GGAGAAG ((((((((((((....((((((((.(((..(....)..)))((..-((((....))))..))........)))))))).)))))..)))))))-....... ( -28.60) >DroSim_CAF1 3440 99 - 1 UUUUCGUGCACACAGUUGCACUUUAUGUAAACAUGUGGGCACACG-CACAAGGUUGUGGAUGAAGAAGAAGAAGUGCAUUGUGCAGGCGAAAA-GGAGAAG ((((((((((((....((((((((..........(((....))).-((((....))))............)))))))).)))))..)))))))-....... ( -28.20) >DroEre_CAF1 3098 99 - 1 UUUUCGUGCACACAGUUGCACUUUAUGUAAACAUGUGGGCACAUG-CACAAGGUUGUGGCAGAAGAAGAAGAAGUGCAUUUUACAGGCGAAAA-GGAGAAG (((((((.........((((((((.......(((((....)))))-((((....))))............))))))))........)))))))-....... ( -22.63) >DroYak_CAF1 3488 99 - 1 UUUUCGUGCAUACAGUUGCACUUUAUGUAAACAUGUGGGCACAUG-CACAAGGUUGUGGCAGAAGAAGAAGAAGUGCAGUGUACAGGCGAAAG-AGAGAAG (((((((...((((.(((((((((.......(((((....)))))-((((....))))............)))))))))))))...)))))))-....... ( -27.00) >consensus UUUUCGUGCACACAGUUGCACUUUAUGUAAACAUGUGGGCACACG_CACAAGGUUGUGGAAGAAGAAGAAGAAGUGCAUUGUACAGGCGAAAA_GGAGAAG (((((((...((((((.(((((((..........(((....)))..((((....))))............)))))))))))))...)))))))........ (-13.65 = -15.65 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:00 2006