
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,156,616 – 5,156,725 |
| Length | 109 |
| Max. P | 0.981848 |

| Location | 5,156,616 – 5,156,725 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.45 |
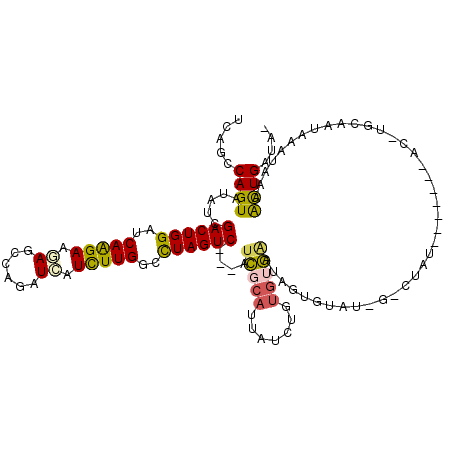
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
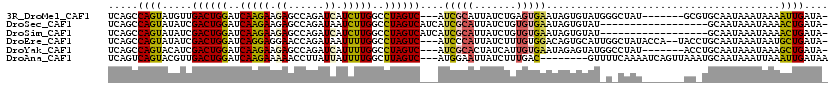
>3R_DroMel_CAF1 5156616 109 - 27905053 UCAGCCAGUAUGUUGACUGGAUCAAGAAGAGCCAGAUCAUCUUGGCCUAGUC---AUCGCAUUAUCUGAGUGAAUAGUGUAUGGGCUAU-------GCGUGCAAUAAAUAAAAUUGAUA- (((..(((.(((((((((((..(((((.((......)).)))))..))))))---)..))))...)))..)))....((((((......-------.))))))................- ( -30.20) >DroSec_CAF1 30518 101 - 1 UCAGCCAGUAUAUCGACUGGAUCAAGAAGAGCCAGAUAAUCUUGGCCUAGUCAUCAUCGCAUUAUCUGUGUGAAUAGUGUAU------------------GCAAUAAAUAAAACUGAUA- .....((((.....((((((..(((((..(......)..)))))..))))))......((((...((((....))))...))------------------))..........))))...- ( -26.20) >DroSim_CAF1 30230 101 - 1 UCAGCCAGUAUAUCGACUGGAUCAAGAAGAGCCAGAUCAUCUUGGCCUAGUCAUCAUCGCAUUAUCUGUGUGAAUAGUGUAU------------------GCAAUAAAUAAAACUGAUA- .....((((.....((((((..(((((.((......)).)))))..))))))......((((...((((....))))...))------------------))..........))))...- ( -29.20) >DroEre_CAF1 29081 114 - 1 UCAGCCAGUAUAUCGACUGGAUCAGGAGGAACCAGAUAAUUUUGGCCUAGUC---AUCCCAUUAUCUUUGUGGACAGUGCAUUGGCUAUACCA--UACCUGCAAUAAAUAAUGCUGAUA- (((((((((......))))...(((((((..(((((....))))))))((((---(...((((.(((....))).))))...)))))......--..))))...........)))))..- ( -29.20) >DroYak_CAF1 28228 109 - 1 UCAGCCAGUACAUCGACUGGAUCAAGAAGAGCCAGAUCAUUUUGGCCUAGUC---AUCGCACUAUCAUUGUGAAUAGAGUAUGGCCUAU-------ACCUGCAAUAAAUAAAGCUGAUA- (((((..((((.((((((((..(((((.((......)).)))))..))))))---.(((((.......)))))...)))))).((....-------....))..........)))))..- ( -28.80) >DroAna_CAF1 52703 109 - 1 UCAGUCAGUACGUUGACUGGAUCAAGAAAAACCUUAUUAUUUUGGCUUAGUC---AUGGAAUUAUCUUUGAC--------GUUUUCAAAAUCAGUUAAAUGCAAUAAAUUAAAUUGAUAA ((((((((....)))))))).....(((((.((..........))....(((---(.(((....))).))))--------.)))))...(((((((.(((.......))).))))))).. ( -21.60) >consensus UCAGCCAGUAUAUCGACUGGAUCAAGAAGAGCCAGAUCAUCUUGGCCUAGUC___AUCGCAUUAUCUGUGUGAAUAGUGUAU_G_CUAU_______AC_UGCAAUAAAUAAAACUGAUA_ .....((((.....((((((..(((((.((......)).)))))..))))))....(((((.......))))).......................................)))).... (-17.59 = -17.23 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:56 2006