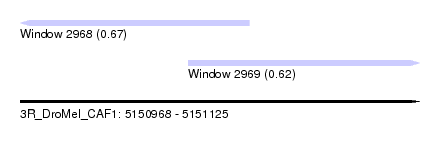
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,150,968 – 5,151,125 |
| Length | 157 |
| Max. P | 0.670040 |

| Location | 5,150,968 – 5,151,058 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.47 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
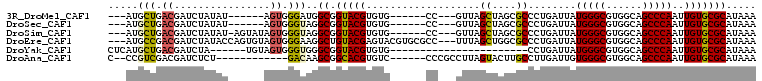
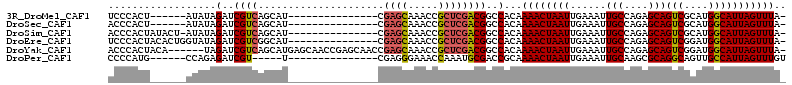
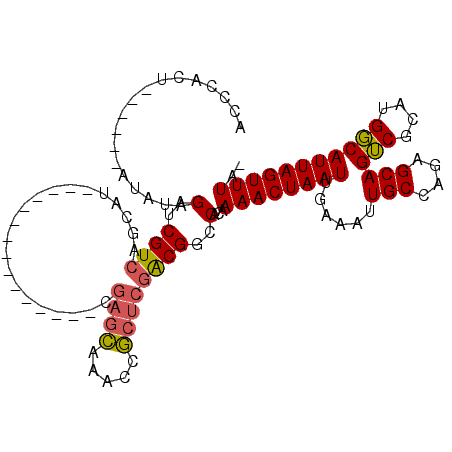
>3R_DroMel_CAF1 5150968 90 - 27905053 ---AUGCUGACGAUCUAUAU------AGUGGGAUGGCGGUACGUGUG------CC---GUUAGCUAGCGCCCUGAUUAUGGGCGUGGCAGCCCAAUUGUGCGCAUAAA ---((((..(((((......------..((((.((((((((....))------))---))))((((.(((((.......)))))))))..)))))))))..))))... ( -35.50) >DroSec_CAF1 24935 90 - 1 ---AUGCUGACGAUCUAUAU------AGUGGGUAGGCGGUACGUGUG------CC---GUUAGCUAGCGCCCUGAUUAUGGGCGUGGCAGCCCAAUUGUGCGCAUAAA ---((((..(((((......------..(((((.(((((((....))------))---))).((((.(((((.......))))))))).))))))))))..))))... ( -36.10) >DroSim_CAF1 24412 95 - 1 ---AUGCUGACGAUCUAUAU-AGUAUAGUGGGUAGGCGGUACGUGUG------CC---GUUAGCUAGCGCCCUGAUUAUGGGCGUGGCAGCCCAAUUGUGCGCAUAAA ---((((.(.....).....-.(((((((((((.(((((((....))------))---))).((((.(((((.......))))))))).)))).)))))))))))... ( -38.40) >DroEre_CAF1 23416 102 - 1 ---AUGCCGACGAUCUAUACCAGUGUAGUGGGAAGGCUGUACGAGUACGUGCGCC---UUUAGCUGGCGCCCUGAUUAUGGGCGUGGCAGCCCAAUUGUGCGCAUAAA ---(((((.((((((((((....)))))(((((((((.(((((....))))))))---))).((((((((((.......))))))..))))))))))))).))))... ( -41.30) >DroYak_CAF1 22521 79 - 1 CUCAUGCUGACGAUCUA------UGUAGUGGGUGGGCGGUACGUGUG-----------------------CCUGAUUAUGGGCGUGGCAGCCCAAUUGUGCGCAUAAA ....(((((.(.(((((------(...)))))).).))))).(((((-----------------------(.((((..(((((......))))))))).))))))... ( -26.50) >DroAna_CAF1 44953 88 - 1 C--CCGUCGACGAUCUCU------------GACAAGCGGCACGUGUC------CCCGCCUUAGUACUUGCCUUGAUUGUGGGCGUGGCAGCCCAAUUGUGCGCAUAAA (--(((((.((((((.((------------....)).((((.(((..------..........))).))))..)))))).)))).))..((.((....)).))..... ( -21.70) >consensus ___AUGCUGACGAUCUAUAU______AGUGGGUAGGCGGUACGUGUG______CC___GUUAGCUAGCGCCCUGAUUAUGGGCGUGGCAGCCCAAUUGUGCGCAUAAA .....(((.((................)).)))..((.(((((...................((....))........(((((......)))))..)))))))..... (-16.55 = -16.47 + -0.08)
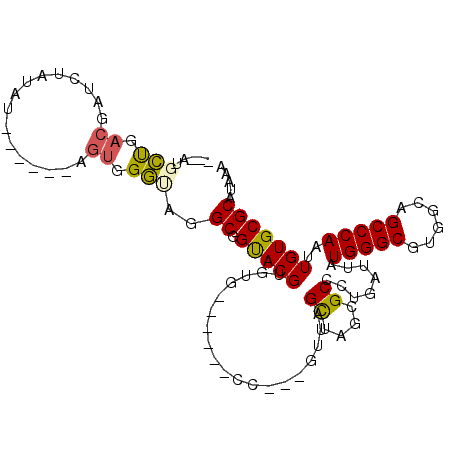
| Location | 5,151,034 – 5,151,125 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -15.06 |
| Energy contribution | -15.04 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5151034 91 + 27905053 UCCCACU------AUAUAGAUCGUCAGCAU---------------CGAGCAAACCGCUCGACGGCCACAAAACUAAUUGAAAUUGCCAGAGCAGUCGCAUGGCAUUAGUUUA- .......------.........((..((.(---------------(((((.....))))))..)).)).(((((((.......(((((..((....)).)))))))))))).- ( -22.41) >DroSec_CAF1 25001 91 + 1 ACCCACU------AUAUAGAUCGUCAGCAU---------------CGAGCAAACCGCUCGACGGCCACAAAACUAAUUGAAAUUGCCAGAGCAGUCGCAUGGCAUUAGUUUA- .......------.........((..((.(---------------(((((.....))))))..)).)).(((((((.......(((((..((....)).)))))))))))).- ( -22.41) >DroSim_CAF1 24478 96 + 1 ACCCACUAUACU-AUAUAGAUCGUCAGCAU---------------CGAGCAAACCGCUCGACGGCCACAAAACUAAUUGAAAUUGCCAGAGCAGUCGCAUGGCAUUAGUUUA- .....(((((..-.)))))...((..((.(---------------(((((.....))))))..)).)).(((((((.......(((((..((....)).)))))))))))).- ( -23.61) >DroEre_CAF1 23488 97 + 1 UCCCACUACACUGGUAUAGAUCGUCGGCAU---------------CGAGCAAACCGCUCGACGGCCACAAAACUAAUUGAAAUUGCCAGAGCAGUCGGAUGGCAUUAGUUUA- (((.(((.(.((((((.........(((.(---------------(((((.....))))))..))).(((......)))....)))))).).))).))).............- ( -28.60) >DroYak_CAF1 22573 106 + 1 ACCCACUACA------UAGAUCGUCAGCAUGAGCAACCGAGCAACCGAGCAAACCGCUCGACGGCCACAAAACUAAUUGAAAUUGCCAGAGCAGUCGGAUGGCAUUAGUUUA- ..........------..(..((((.((.((......)).))....((((.....))))))))..)...(((((((.......(((((...(.....).)))))))))))).- ( -24.11) >DroPer_CAF1 28069 87 + 1 CCCCAUG------CCAGAGAUCGU-----U---------------CGAGGGAAACCAAAUGCGACCGCAAAACUAAUUGAAAUUGCAAGCGCAGGCAGUUGCCAUUAGUUUGU .....((------(....(.((((-----.---------------...((....))....)))).))))((((((((.(.((((((........)))))).).)))))))).. ( -21.40) >consensus ACCCACU______AUAUAGAUCGUCAGCAU_______________CGAGCAAACCGCUCGACGGCCACAAAACUAAUUGAAAUUGCCAGAGCAGUCGCAUGGCAUUAGUUUA_ ..................(..((((.....................((((.....))))))))..)...((((((((......(((....)))(((....))))))))))).. (-15.06 = -15.04 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:54 2006