| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,146,234 – 5,146,331 |
| Length | 97 |
| Max. P | 0.630701 |

| Location | 5,146,234 – 5,146,331 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -16.54 |
| Energy contribution | -18.77 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
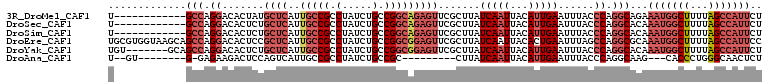
>3R_DroMel_CAF1 5146234 97 + 27905053 U------------GCCAGGACACUAUGCUCAUUGCCGCCUAUCUGCCGGCAGAGUUCGCUUAUCAAUUACAUUGAAUUUACCCAGGCAGAAAUGGCUUUUAGCCAUUCU (------------(((.((.......((((..(((((.(.....).))))))))).......(((((...)))))......)).))))(((.((((.....))))))). ( -26.60) >DroSec_CAF1 20286 97 + 1 U------------GCCAGGACACUCUGCUCAUUGCCGCCUAUCUGCCGGCAGAGUUCGCUUAUCAAUUACAUUGAAUUUACCCAGGCACAAAUGGCUUUUAGCCAUUCU (------------(((.((.......((((..(((((.(.....).))))))))).......(((((...)))))......)).))))..((((((.....)))))).. ( -26.20) >DroSim_CAF1 17271 97 + 1 U------------GCCAGGACACUCUGCUCAUUGCCGCCUAUCUGCCGGCAGAGUUCGCUUAUCAAUUACAUUGAAUUUACCCAGGCACAAAUGGCUUUUAGCCAUUCU (------------(((.((.......((((..(((((.(.....).))))))))).......(((((...)))))......)).))))..((((((.....)))))).. ( -26.20) >DroEre_CAF1 18702 109 + 1 UGCGUGGUAAGCAGCCAGGACACUCCGCUCAUUGCCGCCUAUCUGCCGGCGGAGUUCGCUUAUCAAUUACACUGAAUUUAGCCAGGCGCAAAUGGCUUUUAGCCAUUCC ((((((((((((.....((.....))((((...((((.(.....).)))).))))..))))))).......(((........))))))))((((((.....)))))).. ( -37.20) >DroYak_CAF1 17669 102 + 1 UGU-------GCAGCCAGGACACUCUGCUCAUUGCCGCCUAUCUGCCGGCGGAGUUCGCUUAUCAAUUACAUUGAAUUUACCCAGGCACAAAUGGCUUUUAGCCAUUCU (((-------(((((((((....)))).......(((((........))))).....)))..(((((...)))))..........)))))((((((.....)))))).. ( -27.60) >DroAna_CAF1 40151 86 + 1 U--GU--------G-GAGAAGACUCCAGUCAUUGCCGCCUAUCUGCCGC---------CUUAUCAAUUACAUUGAAUUUACCCAGGCAAG---CACCCUGGGCAACUCU .--.(--------(-(((....))))).........(((((..(((.((---------((....((((......)))).....))))..)---))...)))))...... ( -19.80) >consensus U____________GCCAGGACACUCUGCUCAUUGCCGCCUAUCUGCCGGCAGAGUUCGCUUAUCAAUUACAUUGAAUUUACCCAGGCACAAAUGGCUUUUAGCCAUUCU .............(((.((.......((((..(((((.(.....).))))))))).......(((((...)))))......)).)))...(((((((...))))))).. (-16.54 = -18.77 + 2.22)
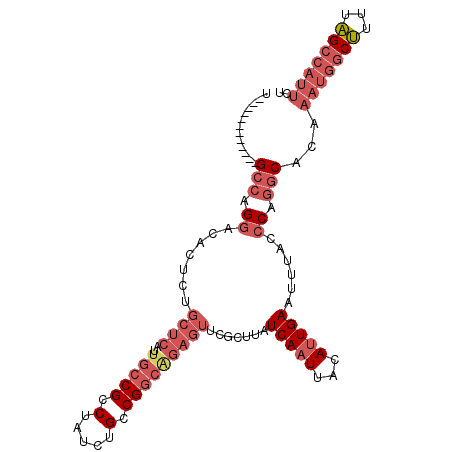
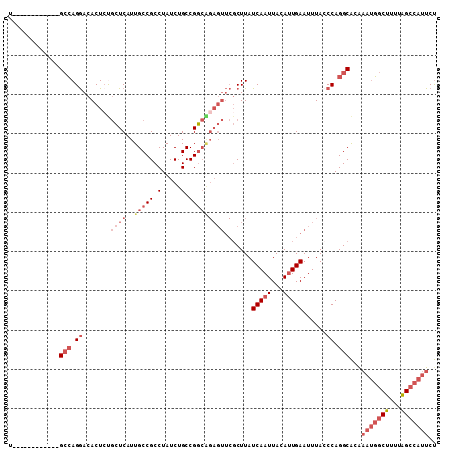
| Location | 5,146,234 – 5,146,331 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5146234 97 - 27905053 AGAAUGGCUAAAAGCCAUUUCUGCCUGGGUAAAUUCAAUGUAAUUGAUAAGCGAACUCUGCCGGCAGAUAGGCGGCAAUGAGCAUAGUGUCCUGGC------------A .(((((((.....)))))))..(((.((..(...(((((...)))))...((...(..(((((.(.....).)))))..).))....)..)).)))------------. ( -30.00) >DroSec_CAF1 20286 97 - 1 AGAAUGGCUAAAAGCCAUUUGUGCCUGGGUAAAUUCAAUGUAAUUGAUAAGCGAACUCUGCCGGCAGAUAGGCGGCAAUGAGCAGAGUGUCCUGGC------------A .(((((((.....))))))).((((.(((...((((.......(((.....))).((((((((.(.....).)))))..)))..))))..))))))------------) ( -31.00) >DroSim_CAF1 17271 97 - 1 AGAAUGGCUAAAAGCCAUUUGUGCCUGGGUAAAUUCAAUGUAAUUGAUAAGCGAACUCUGCCGGCAGAUAGGCGGCAAUGAGCAGAGUGUCCUGGC------------A .(((((((.....))))))).((((.(((...((((.......(((.....))).((((((((.(.....).)))))..)))..))))..))))))------------) ( -31.00) >DroEre_CAF1 18702 109 - 1 GGAAUGGCUAAAAGCCAUUUGCGCCUGGCUAAAUUCAGUGUAAUUGAUAAGCGAACUCCGCCGGCAGAUAGGCGGCAAUGAGCGGAGUGUCCUGGCUGCUUACCACGCA .(((((((.....)))))))..((.(((......(((((...)))))((((((.((((((((........))).((.....)))))))((....))))))))))).)). ( -34.60) >DroYak_CAF1 17669 102 - 1 AGAAUGGCUAAAAGCCAUUUGUGCCUGGGUAAAUUCAAUGUAAUUGAUAAGCGAACUCCGCCGGCAGAUAGGCGGCAAUGAGCAGAGUGUCCUGGCUGC-------ACA .(((((((.....)))))))(((((..((...((((..(((.....))).((.....(((((........)))))......)).))))..))..)..))-------)). ( -33.60) >DroAna_CAF1 40151 86 - 1 AGAGUUGCCCAGGGUG---CUUGCCUGGGUAAAUUCAAUGUAAUUGAUAAG---------GCGGCAGAUAGGCGGCAAUGACUGGAGUCUUCUC-C--------AC--A .((((((((((((...---....))))))).)))))...............---------((.((......)).))......(((((....)))-)--------).--. ( -28.30) >consensus AGAAUGGCUAAAAGCCAUUUGUGCCUGGGUAAAUUCAAUGUAAUUGAUAAGCGAACUCUGCCGGCAGAUAGGCGGCAAUGAGCAGAGUGUCCUGGC____________A .((((((((...))))))))..(((.((..(...(((((...)))))...((.....(((((........)))))......))....)..)).)))............. (-20.71 = -20.97 + 0.26)

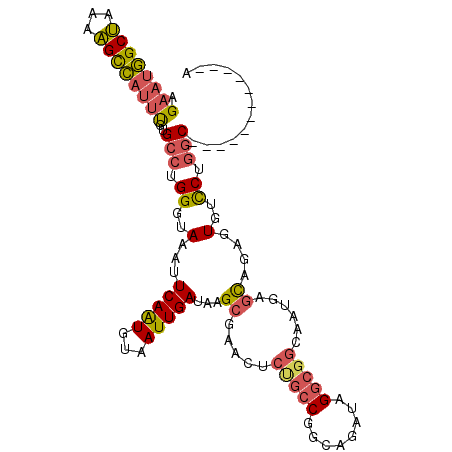
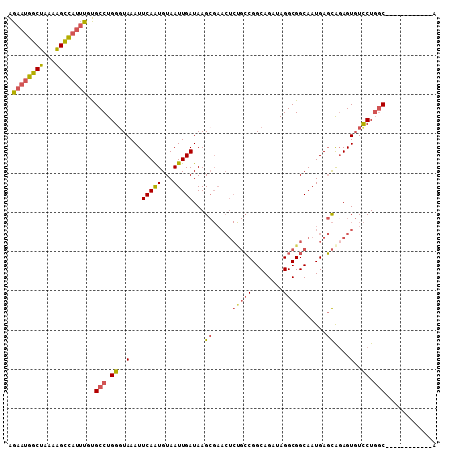
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:52 2006