| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 189,415 – 189,529 |
| Length | 114 |
| Max. P | 0.833989 |

| Location | 189,415 – 189,529 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -43.23 |
| Consensus MFE | -30.72 |
| Energy contribution | -30.12 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.76 |
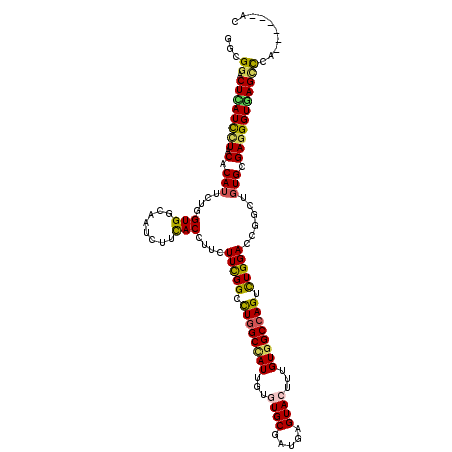
| Structure conservation index | 0.71 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.833989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 189415 114 + 27905053 GGCGGCCUUAUCCUCCACAUACUGGUGGCAGUCUUCACUUUCUUCGGGUUGGCAAUAGUUUGCGAUGAGUACUUUGUGGCCAGUCUGGAUCGGCUUUGCGAGGGUGAGUCCU------CA .(.((.(((((((((..((.((((((.(((((.(((((..((....))..)((((....))))..)))).))..))).)))))).)).....((...))))))))))).)).------). ( -39.00) >DroVir_CAF1 24555 114 + 1 GGUGGACUCAUCCUGCACAUUCUGGUGGCCAUCUAUACGUUCUUCGGCCUGGCUAUUGUGUGCGAUGAGUACUUUGUGGCCAGUCUGGACCGGUUGUGCGAGGGUGAGCCGA------AC ..(((.(((((((((((((..(((((((((...............))))(((((((...((((.....))))...)))))))......))))).))))).))))))))))).------.. ( -51.26) >DroGri_CAF1 23621 114 + 1 GGUGGACUCAUUAUACACAUUCUGGUGGCCAUUUUCACGUUCUUUGGCCUGGCCAUUGUGUGCGAUGAGUACUUUGUGGCCAGUCUGGACAGGCUGUGCGAGGGUGAGCUCG------GC .((.((((((((...((((..(((((((......))))....((..(.((((((((...((((.....))))...)))))))).)..)))))..))))....)))))).)).------)) ( -43.70) >DroWil_CAF1 36909 114 + 1 GGCGGCCUUAUUCUCCACAUCCUGGUGGCAGUUUUCACUUUCUUCGGCCUCGCCAUUGUGUGCGAUGAGUACUUUGUGGCCAGCUUGGAUCGCUUGUGCGAGGGUAAGUCAA------UU ...(((.(((((((((((((((.((((((.................)))..(((((...((((.....))))...)))))..))).))).....)))).)))))))))))..------.. ( -34.93) >DroMoj_CAF1 26507 114 + 1 GGUGGACUCAUCCUACACAUUAUCGUGGCCAUCUAUACCUUCUUUGGCCUGGCCAUUGUGUGCGACGAGUACUUUGUGGCCAGUUUGGACCGGCUGUGCGAGGGUGAGCUCU------AC .(((((((((((((.(.(((..(((.(((((.............)))))(((((((...((((.....))))...)))))))........)))..))).))))))))).)))------)) ( -46.62) >DroAna_CAF1 22981 120 + 1 GGCGGCCUCAUCUUGCACAUCCUGGUGGCAGUCUUCACCUUCUUCGGGCUGGCCAUCGUCUGCGACGAGUACUUUGUGGCCAGUCUGGAUCGACUGUGCGAGGGUAAGAUAAUGAUUUCA ....(((....((((((((((..(((((......)))))...((((((((((((((.(..(((.....)))..).))))))))))))))..)).)))))))))))............... ( -43.90) >consensus GGCGGACUCAUCCUACACAUUCUGGUGGCAAUCUUCACCUUCUUCGGCCUGGCCAUUGUGUGCGAUGAGUACUUUGUGGCCAGUCUGGACCGGCUGUGCGAGGGUGAGCCCA______AC ...((.((((((((.(.(((....(((........)))....(((((.((((((((...((((.....))))...)))))))).)))))......))).))))))))))).......... (-30.72 = -30.12 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:21 2006