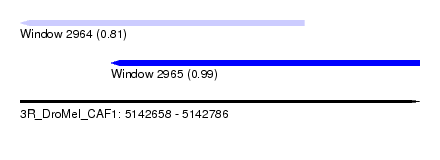
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,142,658 – 5,142,786 |
| Length | 128 |
| Max. P | 0.985419 |

| Location | 5,142,658 – 5,142,749 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -10.40 |
| Energy contribution | -11.30 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5142658 91 - 27905053 GUAU--------CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG-CGAAUU-GGUCGUAAUAAAACGCAUAAAAAGGAUACCGG-CAAAUGGGGA ...(--------((((((.....)))).(((..(((((((((((((..(((-((..((-(.......)))..))))).)))))))..))))-))..)))))) ( -31.40) >DroSec_CAF1 17022 91 - 1 GUAU--------CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG-CGAAUU-GGUCGUAAUAAAACGCAUAAAAAGGAUACCGG-CAAAUGGGGA ...(--------((((((.....)))).(((..(((((((((((((..(((-((..((-(.......)))..))))).)))))))..))))-))..)))))) ( -31.40) >DroSim_CAF1 14046 92 - 1 GUAU--------CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG-CGAAUU-GGUCGUAAUAAAACGCAUAAAAAGGAUACCGGGGAAAUGGGGA ...(--------((((.((((((.((((((((.(((..((((....)))))-))..))-))))))................)))))).)))))......... ( -29.33) >DroEre_CAF1 15294 88 - 1 -----------UCCUGUUAUCCUGGGGGCCAGAUGCUGGUCCUUUUGGAUG-GCAAUU-GUACGUAAUAAAACGCAUAAAAUGGAUCCCGG-CAAAUGGGAA -----------(((((((((((.(((((((((...)))))))))..)))))-))).((-((.(((......))).))))...)))(((((.-....))))). ( -34.20) >DroYak_CAF1 14374 99 - 1 GUAUGCUGUUAUCCUAUUGUCCUGGGGGCCAGAUGCUGGUCCUU-UGGAUGUGGAAUU-GCACGUAAUAAAACGCAUAAAAAGGAUCCAGG-CAAAUGGGGA ...........(((((((..((((((.(((((...)))))((((-(..(((((..(((-(....))))....)))))..))))).))))))-..))))))). ( -32.30) >DroAna_CAF1 36975 81 - 1 GAA------------------AUAAAGGCCACAUGGUGGUCGGAGAAGAUG-GCUCUUAGCCCAUAAUAAAACGCAUAAAAAGGAUCUGUG-AAAAUG-GGA ...------------------.....((((((...))))))((((......-.))))...(((((.......((((...........))))-...)))-)). ( -15.30) >consensus GUAU________CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG_CGAAUU_GGUCGUAAUAAAACGCAUAAAAAGGAUACCGG_CAAAUGGGGA ............(((...((((.((.((((((...))))))))...))))............(((......))).......................))).. (-10.40 = -11.30 + 0.90)

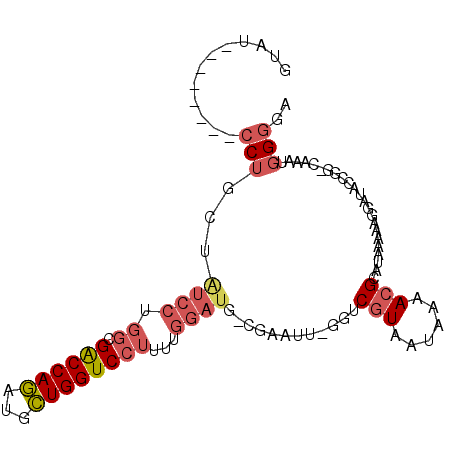
| Location | 5,142,687 – 5,142,786 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.41 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
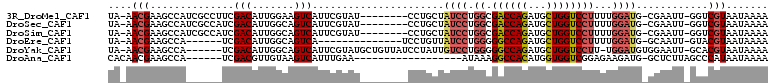
>3R_DroMel_CAF1 5142687 99 - 27905053 UA-AACGAAGCCAUCGCCUUCGACAUUGGAAGUCAUUCGUAU--------CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG-CGAAUU-GGUCGUAAUAAAA ..-.(((..((((..(.((((.......)))).)((((((((--------((.(((.....)))((((((...)))))).....)))))-))))))-))))))....... ( -31.30) >DroSec_CAF1 17051 99 - 1 UA-AACGAAGCCAUCGCCAUCGACAUUGGCAGUCAUUCGUAU--------CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG-CGAAUU-GGUCGUAAUAAAA ..-.(((..((((..((((.......))))....((((((((--------((.(((.....)))((((((...)))))).....)))))-))))))-))))))....... ( -34.60) >DroSim_CAF1 14076 99 - 1 UA-AACGAAGCCAUCGCCAUCGACAUUGGCAGUCAUUCGUAU--------CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG-CGAAUU-GGUCGUAAUAAAA ..-.(((..((((..((((.......))))....((((((((--------((.(((.....)))((((((...)))))).....)))))-))))))-))))))....... ( -34.60) >DroEre_CAF1 15323 87 - 1 UA-AACGAAGCCA------UCGACAUUGGCAGUCA--------------UCCUGUUAUCCUGGGGGCCAGAUGCUGGUCCUUUUGGAUG-GCAAUU-GUACGUAAUAAAA ..-.(((..((((------.......)))).....--------------...((((((((.(((((((((...)))))))))..)))))-)))...-...)))....... ( -31.00) >DroYak_CAF1 14403 101 - 1 UA-AACGAAGCCA------UCGACAUUGGCAGUCAUUCGUAUGCUGUUAUCCUAUUGUCCUGGGGGCCAGAUGCUGGUCCUU-UGGAUGUGGAAUU-GCACGUAAUAAAA ..-.(((((((((------.......)))).....))))).((((((..(((...(((((..((((((((...)))))))).-.))))).)))...-))).)))...... ( -33.40) >DroAna_CAF1 37003 85 - 1 CACAACGAAGCCA------UCGACGUUGUAAGUCAUUUGAA------------------AUAAAGGCCACAUGGUGGUCGGAGAAGAUG-GCUCUUAGCCCAUAAUAAAA ......((.((((------(((((.......))).......------------------.....((((((...))))))......))))-))))................ ( -22.50) >consensus UA_AACGAAGCCA______UCGACAUUGGCAGUCAUUCGUAU________CCUGCUAUCCUGGCGACCAGAUGCUGGUCCUUUUGGAUG_CGAAUU_GGUCGUAAUAAAA ....(((..............(((.......)))......................((((.((.((((((...))))))))...))))............)))....... (-13.44 = -13.97 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:50 2006