| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,117,653 – 5,117,811 |
| Length | 158 |
| Max. P | 0.877614 |

| Location | 5,117,653 – 5,117,771 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -23.88 |
| Energy contribution | -23.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5117653 118 + 27905053 GUGUUUGUUUGUUUGC--UGCCUUUUGUUCGCUUGUAGUGGUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGACACGUUGAAGUAUGAAAGGAAUUAACAAAUACU (((((((((.(((...--..(((((..(..((((...(((...((((..(((((.(((.....))).)))))..........)))).)))....)))))..)))))))).))))))))). ( -26.20) >DroSec_CAF1 19250 120 + 1 GUGUUUGUUUGUUUGCUUUGCCUUUUGUUCGCUUGUAGUGGUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAGGAAUUAACAAAUACU (((((((((.(((.......(((((..(..((((...(((((..(((..(((((.(((.....))).)))))....)))......)))))....)))))..)))))))).))))))))). ( -28.61) >DroSim_CAF1 19825 118 + 1 GUGUUUGUUUGUUUGC--UGCCUUUUGUUCGCUUGUAGUGGUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAGGAAUUAACAAAUACU (((((((((.(((...--..(((((..(..((((...(((((..(((..(((((.(((.....))).)))))....)))......)))))....)))))..)))))))).))))))))). ( -29.20) >DroEre_CAF1 20725 115 + 1 -UG--GGUGUGUUUGC--UGCUUUUUGUUCGCUUGUAGUGGUGCUGAUUGUGUUUGCGAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAAGAAUUAACAAAUACU -..--...(((((((.--..(((((..(..((((...(((((..(((..(((((.((((...)))).)))))....)))......)))))....)))))..)))))......))))))). ( -24.50) >DroYak_CAF1 20698 116 + 1 GUG--GGUGUGUUUGC--UGCCUUUUGUUCGCUUGUAGUGGUGCUGAUUGUGUUUGCGAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGCAUGAAAGGCAUUAACAAAUACU ...--...(((((((.--(((((((..(..((((...(((((..(((..(((((.((((...)))).)))))....)))......)))))....)))))..)))))))....))))))). ( -32.60) >consensus GUGUUUGUUUGUUUGC__UGCCUUUUGUUCGCUUGUAGUGGUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAGGAAUUAACAAAUACU ......(((((((.......(((((..(..((((...(((((..(((..(((((.((((...)))).)))))....)))......)))))....)))))..)))))....)))))))... (-23.88 = -23.76 + -0.12)

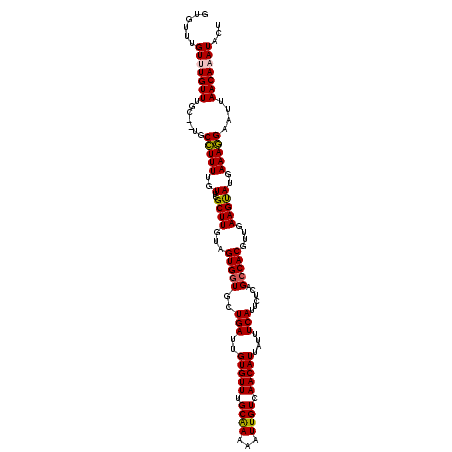

| Location | 5,117,691 – 5,117,811 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.56 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5117691 120 + 27905053 GUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGACACGUUGAAGUAUGAAAGGAAUUAACAAAUACUUACAGCAGACAUUGCCUGCAGAAUUUUAGUUUUUGCAAAU (((((((..(((((.(((.....))).)))))..........)))).)))((((((((((...............)))))).))))..........(((((((.......)))))))... ( -23.86) >DroSec_CAF1 19290 120 + 1 GUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAGGAAUUAACAAAUACUUACAGCAGACAUUGCCUGCAGAAUUUUAGUUUUUGUAAAU .(((((...(((((((.......((....))...((((((.((((((...)))).)).))))))........)))))))...))))).........(((((((.......)))))))... ( -23.50) >DroSim_CAF1 19863 120 + 1 GUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAGGAAUUAACAAAUACUUACAGCAGACAUUGCCUGCAGAAUUUUAGUUUUUGCAAAA .(((((...(((((((.......((....))...((((((.((((((...)))).)).))))))........)))))))...))))).........(((((((.......)))))))... ( -25.50) >DroEre_CAF1 20760 120 + 1 GUGCUGAUUGUGUUUGCGAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAAGAAUUAACAAAUACUUACAGCAAACAUGGCCUGCACAAUUUUAGUUAUUGCACAU ((((.(.(..(((((((((......)).......((((((.((((((...)))).)).))))))....................)))))))..).).))))................... ( -25.70) >DroYak_CAF1 20734 120 + 1 GUGCUGAUUGUGUUUGCGAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGCAUGAAAGGCAUUAACAAAUACUUACAGCAAACAUUGCCUGCAGAAGUUUAGUUUUUGCACAU .(((((...(((((((..((...((((.......((((((.((((((...)))).)).))))))))))))..)))))))...))))).........((((((((.....))))))))... ( -26.71) >consensus GUGCUGAUUGUGUUUGCAAAAAUUGUCAACAUUAUUUCAUUCUCAGCCACGUUGAAGUAUGAAAGGAAUUAACAAAUACUUACAGCAGACAUUGCCUGCAGAAUUUUAGUUUUUGCAAAU .(((((...(((((((.......((....))...((((((.((((((...)))).)).))))))........)))))))...))))).........(((((((.......)))))))... (-23.12 = -23.56 + 0.44)

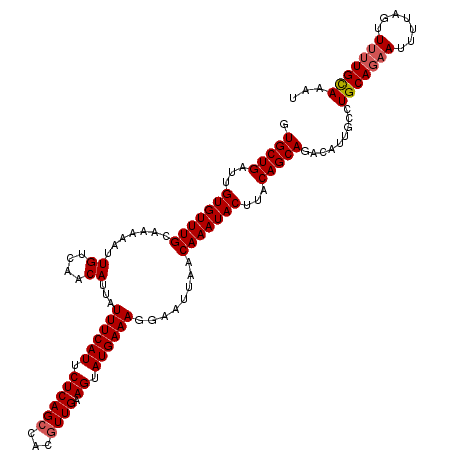

| Location | 5,117,691 – 5,117,811 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -24.74 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.56 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5117691 120 - 27905053 AUUUGCAAAAACUAAAAUUCUGCAGGCAAUGUCUGCUGUAAGUAUUUGUUAAUUCCUUUCAUACUUCAACGUGUCUGAGAAUGAAAUAAUGUUGACAAUUUUUGCAAACACAAUCAGCAC .((((((((((..........((((((...)))))).........((((((((...((((((.((.((.......)))).))))))....))))))))))))))))))............ ( -26.50) >DroSec_CAF1 19290 120 - 1 AUUUACAAAAACUAAAAUUCUGCAGGCAAUGUCUGCUGUAAGUAUUUGUUAAUUCCUUUCAUACUUCAACGUGGCUGAGAAUGAAAUAAUGUUGACAAUUUUUGCAAACACAAUCAGCAC .....................((((((...))))))((((((...((((((((...((((((.((.((.......)))).))))))....))))))))..)))))).............. ( -22.40) >DroSim_CAF1 19863 120 - 1 UUUUGCAAAAACUAAAAUUCUGCAGGCAAUGUCUGCUGUAAGUAUUUGUUAAUUCCUUUCAUACUUCAACGUGGCUGAGAAUGAAAUAAUGUUGACAAUUUUUGCAAACACAAUCAGCAC .((((((((((..........((((((...)))))).........((((((((...((((((.((.((.......)))).))))))....))))))))))))))))))............ ( -26.40) >DroEre_CAF1 20760 120 - 1 AUGUGCAAUAACUAAAAUUGUGCAGGCCAUGUUUGCUGUAAGUAUUUGUUAAUUCUUUUCAUACUUCAACGUGGCUGAGAAUGAAAUAAUGUUGACAAUUUUCGCAAACACAAUCAGCAC .((..((((.......))))..)).....(((((((.(.(((...((((((((...((((((.((.((.......)))).))))))....)))))))).))))))))))).......... ( -25.00) >DroYak_CAF1 20734 120 - 1 AUGUGCAAAAACUAAACUUCUGCAGGCAAUGUUUGCUGUAAGUAUUUGUUAAUGCCUUUCAUGCUUCAACGUGGCUGAGAAUGAAAUAAUGUUGACAAUUUUCGCAAACACAAUCAGCAC ..((((.........(((((.((((((...)))))).).))))..(((((((((..((((((..((((.......)))).))))))...)))))))))..................)))) ( -23.40) >consensus AUUUGCAAAAACUAAAAUUCUGCAGGCAAUGUCUGCUGUAAGUAUUUGUUAAUUCCUUUCAUACUUCAACGUGGCUGAGAAUGAAAUAAUGUUGACAAUUUUUGCAAACACAAUCAGCAC ...(((...............((((((...))))))((((((...((((((((...((((((..((((.......)))).))))))....))))))))..))))))..........))). (-22.20 = -22.56 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:36 2006