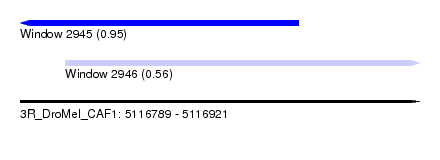
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,116,789 – 5,116,921 |
| Length | 132 |
| Max. P | 0.950007 |

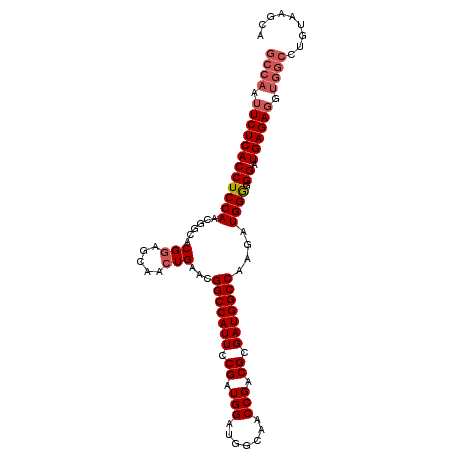
| Location | 5,116,789 – 5,116,881 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -31.95 |
| Energy contribution | -33.73 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5116789 92 - 27905053 GCCAAUUCUCACCUCCAAAGGAACGAAGCAAAUGGGCGGCCAUUCCGAUGGAUGAAAUCCGACGCGAUGGCCACGAUGGA-GGAUGAGA-----------AGCA ((...((((((((((((..(.......((......))(((((((.((.(((((...))))).)).))))))).)..))))-)).)))))-----------))). ( -39.10) >DroSec_CAF1 18396 104 - 1 GCCAAUUCUCACCUCCAACGGCACGGAGCAACUGAACGGCCAUUCCGAUGGAUGGCAGCCGACGCGAUGGCCAAGAUGGGGGGAUGAGAGGUGGCCUGUAAGCA ((((.(((((((((((..((((.(((.....)))....(((((((....))))))).))))..((....)).......))))).)))))).))))......... ( -41.90) >DroSim_CAF1 18958 104 - 1 GCCAAUUCUCACCUCCAACGGCACGGAGCAACUGAACGGCCAUUCCGAUGGAUGGCAACCGACGCGAUGGCCAAGAUGGGGGGAUGAGAGGUGGCCUGUAAGCA ((((.(((((((((((....((.....))........(((((((.((.(((.......))).)).)))))))......))))).)))))).))))......... ( -40.80) >consensus GCCAAUUCUCACCUCCAACGGCACGGAGCAACUGAACGGCCAUUCCGAUGGAUGGCAACCGACGCGAUGGCCAAGAUGGGGGGAUGAGAGGUGGCCUGUAAGCA ((((.((((((((((((......(((.....)))...(((((((.((.(((.......))).)).)))))))....)))).)).)))))).))))......... (-31.95 = -33.73 + 1.78)


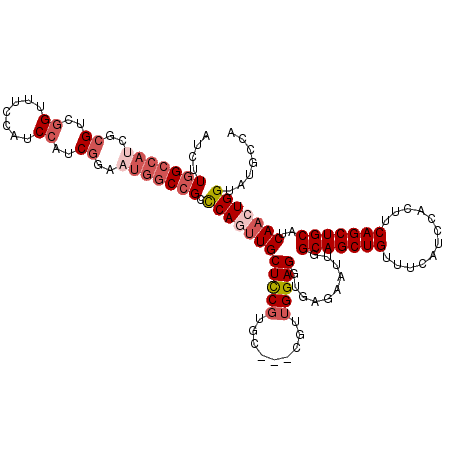
| Location | 5,116,804 – 5,116,921 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -28.85 |
| Energy contribution | -30.90 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5116804 117 + 27905053 AUCGUGGCCAUCGCGUCGGAUUUCAUCCAUCGGAAUGGCCGCCCAUUUGCUUCGUUC---CUUUGGAGGUGAGAAUUGGCAGCUGUUUCAUCCACUUCAGCUGCAUCAACUGGUAUGCCA ...((((((((..((..((((...))))..))..))))))))(((((..((((....---.....))))..)).....(((((((............)))))))......)))....... ( -38.90) >DroSec_CAF1 18423 117 + 1 AUCUUGGCCAUCGCGUCGGCUGCCAUCCAUCGGAAUGGCCGUUCAGUUGCUCCGUGC---CGUUGGAGGUGAGAAUUGGCAGCUGCUUCAUCCACUUCAGCUGCAUCAACUGGUAUGCCA .....((((((..((..((.......))..))..))))))..(((((((....((((---.((((.(((((.((...(((....)))...))))))))))).)))))))))))....... ( -39.20) >DroSim_CAF1 18985 117 + 1 AUCUUGGCCAUCGCGUCGGUUGCCAUCCAUCGGAAUGGCCGUUCAGUUGCUCCGUGC---CGUUGGAGGUGAGAAUUGGCAGCUGUUUCAUCCACUUCAGCUGCAUCAACUGGUAUGCCA .....((((((..((..((.......))..))..))))))..((((((((((((...---...)))))..........(((((((............)))))))..)))))))....... ( -40.10) >DroEre_CAF1 19933 114 + 1 ----UGGCCAUC--GUCCGUAUCCAUCCAUCGGAUUGGCCGCCCAAAUGCUCCGUGCCUCCGUUGGAGGUGAGAAUUGGCAGCUGUUUCAUACACUUCAGCUGCAUCAACUGGUACGCCA ----.(((((..--(((((...........))))))))))((((((...(((...((((((...)))))))))..)))(((((((............))))))).......)))...... ( -40.00) >DroYak_CAF1 19868 111 + 1 AUCUUGGCCAUC--GUCGGUUUCCAUCCAUC-------CAGCCCAGUUGCUCCUUGCCUCCGUUGGAGGUGAGAAUUGGCAGCUGUUUCAUACACUUCAGCUGCAUCAACUGGUAUUCCA .....((((...--...))))..........-------....(((((((...((..(((((...)))))..)).....(((((((............)))))))..)))))))....... ( -36.20) >consensus AUCUUGGCCAUCGCGUCGGUUUCCAUCCAUCGGAAUGGCCGCCCAGUUGCUCCGUGC___CGUUGGAGGUGAGAAUUGGCAGCUGUUUCAUCCACUUCAGCUGCAUCAACUGGUAUGCCA ....(((((((..((..((.......))..))..))))))).((((((((((((.........)))))..........(((((((............)))))))..)))))))....... (-28.85 = -30.90 + 2.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:32 2006