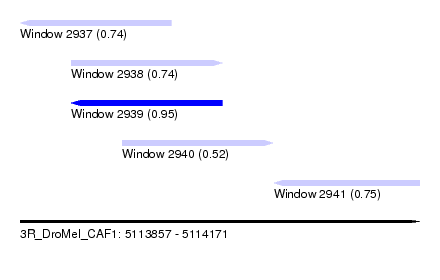
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,113,857 – 5,114,171 |
| Length | 314 |
| Max. P | 0.949863 |

| Location | 5,113,857 – 5,113,976 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -24.62 |
| Energy contribution | -25.46 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5113857 119 - 27905053 CAGACUUACAAUACCAUUUAAUUACAAUCGAUUGC-AAUGAAAACAACAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCUGGCCGCAAACGUUGCCGGCACUUCGAGGUAGUAAACAGAU ....................(((((..((((.(((-(((.((((.((((......))))...)))).))))))....((((((.((....)).))))))...)))).)))))........ ( -28.80) >DroSec_CAF1 15487 119 - 1 CAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC-AUUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCAUGCCGCAAACGUUGCCGGCACUUCGAGGUGGUAAACAGAU .....((((.(((((((.......(((((....))-.)))......(((((((((((...))))))).))))..)))))))((((((.....)).))))...........))))...... ( -29.00) >DroSim_CAF1 16019 119 - 1 CAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC-AAUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCACGCCGCAAACGUUGCCGGCACUUCGAGGUGGCAAACAGAU ...........((((((((......(((.(...((-((((......(((((((((((...))))))).)))).(((((....)))))..))))))...).)))..))))))))....... ( -33.80) >DroEre_CAF1 16849 117 - 1 CGGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC-AAUGAAAACAACG--AAAAUGUUUCAUUUUGAUUGCAUGUGGCUGGCCACACACGUUGCCGGCCCUUUGAGGUAGUAAACAGAU .....((((..((((........(((.(((((((.-(((((((.((...--....))))))))).))))))).)))(((((((.((....)).)))))))......))))))))...... ( -33.60) >DroYak_CAF1 16789 120 - 1 CAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGCCACUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCUGGCCGCAAACGUUGCCGGCACUUUGAGGUAGUAAACAGAU .....((((..((((.......((((.((((((((..(((....)))..))((((((...)))))))))))).))))((((((.((....)).)))))).......))))))))...... ( -31.70) >consensus CAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC_AAUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCUGGCCGCAAACGUUGCCGGCACUUCGAGGUAGUAAACAGAU ...........((((.......((((.((((((...(((((((.((.........)))))))))..)))))).))))((((((.((....)).)))))).......)))).......... (-24.62 = -25.46 + 0.84)

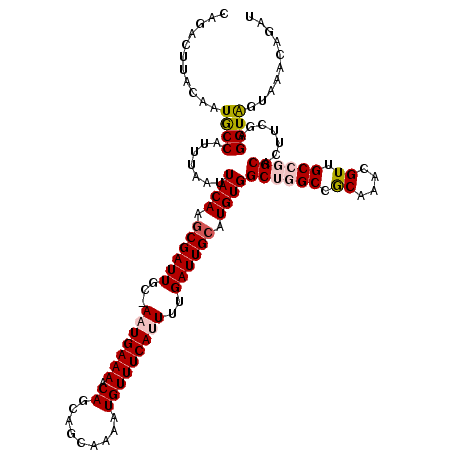
| Location | 5,113,897 – 5,114,016 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.44 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5113897 119 + 27905053 AGCCACAUGCAAUCAAAAUGAAACAUUUUGCUGUUGUUUUCAUU-GCAAUCGAUUGUAAUUAAAUGGUAUUGUAAGUCUGCUCAUUAAAACAACACUCGCACAUGCCAUAUUAAAGGUGA .(((...(((((((((((((...))))))...(((((.......-))))).))))))).(((((((((((((..(((.................)))..)).))))))).)))).))).. ( -23.83) >DroSec_CAF1 15527 119 + 1 UGCCACAUGCAAUCAAAAUGAAACAUUUUGCUGCUGUUUUCAAU-GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUAAAACAACAAUCGCACAUGCCAUAUUAAAGGUUU .((((((.(((..(((((((...))))))).))))))......(-((((....))))).(((((((((((.((..(..((.............))..)..))))))))).)))).))).. ( -20.42) >DroSim_CAF1 16059 119 + 1 UGCCACAUGCAAUCAAAAUGAAACAUUUUGCUGCUGUUUUCAUU-GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUAAAACAACACUUGCACAUGCCAUAUUAAAGGUUU .((((((.(((..(((((((...))))))).))))))....(((-((((....)))))))...((((((((((((((.................))))))).)))))))......))).. ( -27.43) >DroEre_CAF1 16889 117 + 1 AGCCACAUGCAAUCAAAAUGAAACAUUUU--CGUUGUUUUCAUU-GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCCGCUCAUUUAAUCGACACUCGCACAUGCCAUGUUAAAGGCAU .((((((.((.((...(((((((((....--...)).)))))))-...)).)).)))..(((((((((((((..(((....((........)).)))..)).))))))).)))).))).. ( -27.20) >DroYak_CAF1 16829 120 + 1 AGCCACAUGCAAUCAAAAUGAAACAUUUUGCUGCUGUUUUCAGUGGCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUUAAUCGACACUCGCACAUGCCAUAUUAAAGGCAU .((((((.((..((.....))......((((..(((....)))..))))..)).)))..(((((((((((((..(((....((........)).)))..)).))))))).)))).))).. ( -33.30) >consensus AGCCACAUGCAAUCAAAAUGAAACAUUUUGCUGCUGUUUUCAUU_GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUAAAACAACACUCGCACAUGCCAUAUUAAAGGUAU .((((((.((...(((((((...)))))))....(((........)))...)).)))..((((((((((((((.(((.((..........))..))).))).))))))).)))).))).. (-19.64 = -19.44 + -0.20)


| Location | 5,113,897 – 5,114,016 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -24.18 |
| Energy contribution | -25.54 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5113897 119 - 27905053 UCACCUUUAAUAUGGCAUGUGCGAGUGUUGUUUUAAUGAGCAGACUUACAAUACCAUUUAAUUACAAUCGAUUGC-AAUGAAAACAACAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCU .............(.(((((((((.(((((((((..((((....))))......(((((((((......))))).-))))))))))))).(((((((...))))))).))))))))).). ( -26.80) >DroSec_CAF1 15527 119 - 1 AAACCUUUAAUAUGGCAUGUGCGAUUGUUGUUUUAAUGAGCAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC-AUUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCA ............((.(((((((((((((((((((..((((....))))((((((.(((...........))).))-))))))))))))).(((((((...))))))))))))))))).)) ( -31.80) >DroSim_CAF1 16059 119 - 1 AAACCUUUAAUAUGGCAUGUGCAAGUGUUGUUUUAAUGAGCAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC-AAUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCA ......((((.(((((((.((.((((.((((((....)))))))))).)))))))))))))(((((.(((((((.-(((((((.((.........))))))))).))))))).))))).. ( -31.70) >DroEre_CAF1 16889 117 - 1 AUGCCUUUAACAUGGCAUGUGCGAGUGUCGAUUAAAUGAGCGGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC-AAUGAAAACAACG--AAAAUGUUUCAUUUUGAUUGCAUGUGGCU ..(((.((((.(((((((.((..(((.(((.((....)).))))))..)))))))))))))..(((.(((((((.-(((((((.((...--....))))))))).))))))).)))))). ( -32.30) >DroYak_CAF1 16829 120 - 1 AUGCCUUUAAUAUGGCAUGUGCGAGUGUCGAUUAAAUGAGCAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGCCACUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCU ..(((.......(((((..(((...(((.(((((((((.(((.........))))))))))))))).)))..)))))......((((((((((((((...))))))).)))).)))))). ( -38.10) >consensus AAACCUUUAAUAUGGCAUGUGCGAGUGUUGUUUUAAUGAGCAGACUUACAAUGCCAUUUAAUUACAAGCGAUUGC_AAUGAAAACAGCAGCAAAAUGUUUCAUUUUGAUUGCAUGUGGCU ......((((.(((((((.((..(((.((((((....)))))))))..)))))))))))))(((((.((((((...(((((((.((.........)))))))))..)))))).))))).. (-24.18 = -25.54 + 1.36)

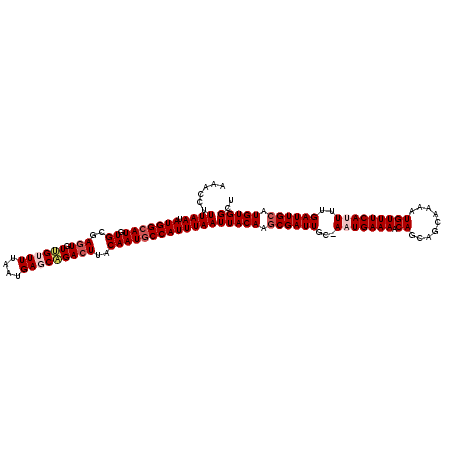
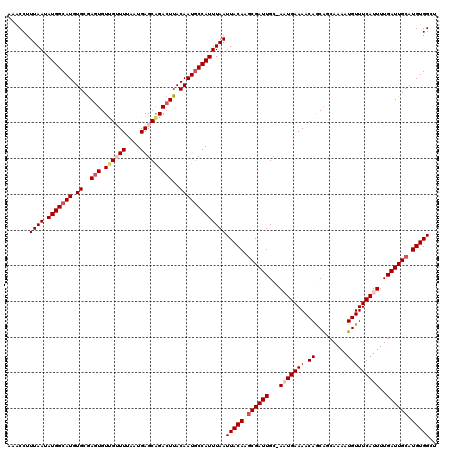
| Location | 5,113,937 – 5,114,056 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5113937 119 + 27905053 CAUU-GCAAUCGAUUGUAAUUAAAUGGUAUUGUAAGUCUGCUCAUUAAAACAACACUCGCACAUGCCAUAUUAAAGGUGAUCAUUGGCCAACAUGCAUACACAAUUUUAGUCCCACUAGC ...(-(((...(((..(..(((((((((((((..(((.................)))..)).))))))).))))..)..)))..((.....)))))).........((((.....)))). ( -19.23) >DroSec_CAF1 15567 119 + 1 CAAU-GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUAAAACAACAAUCGCACAUGCCAUAUUAAAGGUUUUCAUUGGCCAACAAGCAUACACAAUUACUGACCCACUGGC ....-......((((((..(((((((((((.((..(..((.............))..)..))))))))).)))).((((......)))).))))))........................ ( -21.02) >DroSim_CAF1 16099 119 + 1 CAUU-GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUAAAACAACACUUGCACAUGCCAUAUUAAAGGUUUUCAUUGGCCAACAAGCAUACACAAUUACUGACCCACUGGC ....-......((((((..((((((((((((((((((.................))))))).))))))).)))).((((......)))).))))))........................ ( -27.03) >DroEre_CAF1 16927 118 + 1 CAUU-GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCCGCUCAUUUAAUCGACACUCGCACAUGCCAUGUUAAAGGCAUUAUUCGCCC-ACAAGCAUACCCAAUUACCAUCCCAAUAGC ....-......((((((..(((((((((((((..(((....((........)).)))..)).))))))).)))).(((.......))).-))))))........................ ( -24.40) >DroYak_CAF1 16869 120 + 1 CAGUGGCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUUAAUCGACACUCGCACAUGCCAUAUUAAAGGCAUUAUUUGCCCAACAAGCAUACACAAUUACGGUCCCAAUGGC ..(((......((((((..(((((((((((((..(((....((........)).)))..)).))))))).)))).((((.....))))..))))))...))).................. ( -27.80) >consensus CAUU_GCAAUCGCUUGUAAUUAAAUGGCAUUGUAAGUCUGCUCAUUAAAACAACACUCGCACAUGCCAUAUUAAAGGUAUUCAUUGGCCAACAAGCAUACACAAUUACUGUCCCACUGGC ...........((((((..((((((((((((((.(((.((..........))..))).))).))))))).)))).(((........))).))))))........................ (-17.64 = -18.08 + 0.44)

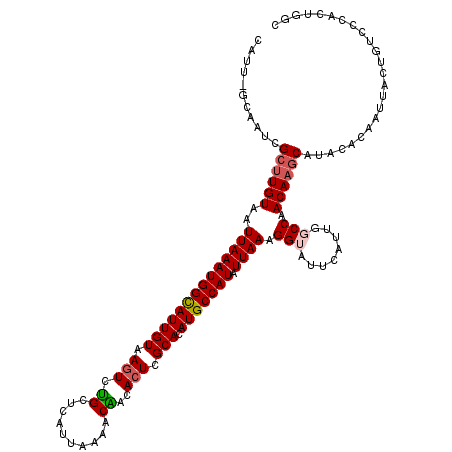
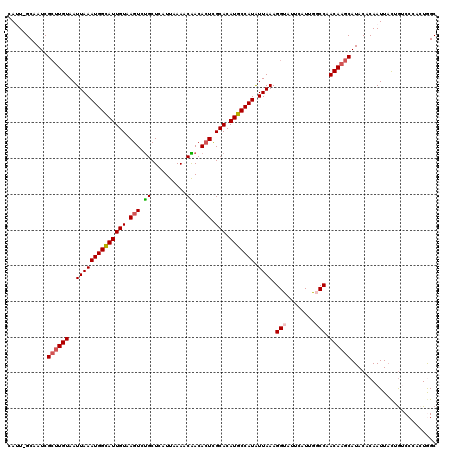
| Location | 5,114,056 – 5,114,171 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.01 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -23.54 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5114056 115 - 27905053 G--CUACUAACUCAAUCUUGACCGUUUGAUUGAUAGAUUGAUUGAUGCAGGGCAGCCGAA-AUUGAAUUUUUGCUCUACGA--AACAAUUGGUUAUGCAUUAAUUGAUUGAAUCAUGGUC .--................(((((..(((((....(((..(((((((((....((((((.-.(((...(((((.....)))--))))))))))).)))))))))..))).)))))))))) ( -27.70) >DroSec_CAF1 15686 118 - 1 U--CUAGUAACUCAAUCUUGACCGUUUGAUUAAUAGAUUGAUUGAUGCAGGUCAGCCGAAAAUUGGAUUUUUGCCCUGCGAGAAGCAAUUGGUUAUGCAUUAAUUGAUUGAAUCAUGGUC .--................(((((..(((((....(((..(((((((((....((((((...(((..(((((((...))))))).))))))))).)))))))))..))).)))))))))) ( -33.70) >DroSim_CAF1 16218 120 - 1 UCGCUACUAACUCAAUCUUGAACGUUUGAUUGAUAGAUUGAUUGAUGCAGGUCAGCCGAAAAUUGGAUUUUUGCUCUGCGAGAAGCAAUUGGUUAUGCAUUAAUUGAUUGAAUCAUGGUC ..((((.....((((((..........))))))..(((..(((((((((....((((((...(((..(((((((...))))))).))))))))).)))))))))..)))......)))). ( -30.50) >DroEre_CAF1 17045 114 - 1 G--CUACUAACUCAAUCUUGAGCGAUUGAUUGCCAGAUUGAUUGAUGCAGGUCAGCCGAA-AUUGAAUUUUUGCCC-UCG--AAGCAAUUGGUUGUGCAUUAAUUGAUUGAAUCAUGCUC .--................(((((..(((((....(((..(((((((((...(((((((.-.........((((..-...--..))))))))))))))))))))..))).)))))))))) ( -29.80) >DroYak_CAF1 16989 115 - 1 G--CUACUAACUCAAUCUUGAGCGAUUGAUUGAUAGAUUGAUUGAUGCAGGGCAGCCAAA-AUUGAAUUUUUGCUCUUCG--AAGCAAUUGGUUGGGCAUUAAUUGAUUGAAUCAUGGUC .--..((((..((((((......)))))).((((.(((..((((((((....(((((((.-.........(((((.....--.)))))))))))).))))))))..)))..)))))))). ( -32.50) >consensus G__CUACUAACUCAAUCUUGACCGUUUGAUUGAUAGAUUGAUUGAUGCAGGUCAGCCGAA_AUUGAAUUUUUGCUCUGCGA_AAGCAAUUGGUUAUGCAUUAAUUGAUUGAAUCAUGGUC ...................(((((..(((((....(((..(((((((((....((((((...........(((((........))))))))))).)))))))))..))).)))))))))) (-23.54 = -24.42 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:27 2006