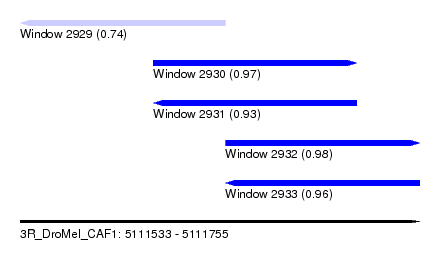
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,111,533 – 5,111,755 |
| Length | 222 |
| Max. P | 0.980341 |

| Location | 5,111,533 – 5,111,647 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.65 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -22.55 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5111533 114 - 27905053 GCUGCAGCAUGCUUUAAGGCGCUUCUUGAUUAGCAAAAGCCCUGCUCUC-GAAUUAGCGUCUAUCUUUAUUUUAGCCUUGCCCGUAGCGCUUUCAUUUCAG-----CAGCCGCAUUUUGU (((((.(((.(((...(((((((..((((..((((.......)))).))-))...)))))))...........)))..)))..)))))(((........))-----)............. ( -27.94) >DroSec_CAF1 13039 114 - 1 AGUGCAGCAUGCUUUAAGUCGCUUCUUGAUUAGCAAAAGCCCUGCUCUC-GAAUUAGCGUCUAUCCUUAUUUUAGCCUUGCCCGCAGCGCUUUCAUUUCAG-----CAGCUGCAUUUUGU ((((((((.((((...((.((((..((((..((((.......)))).))-))...)))).))...........(((.(((....))).)))........))-----)))))))))).... ( -31.40) >DroSim_CAF1 13593 111 - 1 AGUGCAGCAUGCUUUAAGGCGCCUCUUGAUUAGCAAAAGCCCUGCUCUC-GAAUUAGCGUCUAUCCUUAUUUUAGCCUUGCCCGCAGCGCUUUCAUUUCA--------GCUGCAUUUUGU ((((((((........((((((...((((..((((.......)))).))-))....))))))...........(((.(((....))).))).........--------)))))))).... ( -28.30) >DroEre_CAF1 14360 120 - 1 AAUGUAGCAUGCUUUAAGCCGCUUCUUGAUUAGCAAAAGCCGUGCUCUCGAAAUUAGCGUCUAUCUUUAUUUUAGCCUUGCCCGCAGCGCUUUCAUUUCAGCACUGCAGCUGCAUUUUGU ((((((((.(((((((((......)))))..))))...((.(((((...(((((.((((((((.........)))...((....)))))))...)))))))))).)).)))))))).... ( -31.40) >DroYak_CAF1 14324 119 - 1 AAUGUAGCAUGCUUUAAGGCGCUUCUUGAUUAGCCACAGCCGUGCUCUC-AAAUUAGCGUCUAUCUCUAUUUUAGCCUUGCCCGCAGCGCUUUCAUUUCGCCUCUACAGCUGCAUUUUGU ((((((((........(((((((..((((..(((.((....))))).))-))...)))))))...........(((.(((....))).))).................)))))))).... ( -28.80) >consensus AAUGCAGCAUGCUUUAAGGCGCUUCUUGAUUAGCAAAAGCCCUGCUCUC_GAAUUAGCGUCUAUCUUUAUUUUAGCCUUGCCCGCAGCGCUUUCAUUUCAG_____CAGCUGCAUUUUGU ((((((((........(((((((........((((.......)))).........)))))))...........(((.(((....))).))).................)))))))).... (-22.55 = -23.03 + 0.48)



| Location | 5,111,607 – 5,111,720 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -24.91 |
| Energy contribution | -26.95 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5111607 113 + 27905053 GCUUUUGCUAAUCAAGAAGCGCCUUAAAGCAUGCUGCAGCUUU-------UGUUGCAACAUGUACUUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUAC ((((((........))))))........(((((.((((((...-------.)))))).)))))(((((((.((((((.(((((((((.....))))))).)).)))))).)))))))... ( -42.50) >DroSec_CAF1 13113 113 + 1 GCUUUUGCUAAUCAAGAAGCGACUUAAAGCAUGCUGCACUUUU-------UGUUGCAACAUGUACCUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGAAAAAGCAAAGUUAC ((((((........))))))(((((...(((((.(((((....-------.).)))).)))))....(((.((((((.(((((((((.....))))))).)).)))))).)))))))).. ( -35.80) >DroSim_CAF1 13664 112 + 1 GCUUUUGCUAAUCAAGAGGCGCCUUAAAGCAUGCUGCACUU-U-------UGUUGCAACAUGUACCUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGAAAAAGCAAAGUUAC ((((((........))))))........(((((.(((((..-.-------.).)))).)))))((.((((.((((((.(((((((((.....))))))).)).)))))).)))).))... ( -33.90) >DroEre_CAF1 14440 120 + 1 GCUUUUGCUAAUCAAGAAGCGGCUUAAAGCAUGCUACAUUUUUUGUGUUUUUUACCAACAUGUAUUUCGCAUUUUUCGGCACUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUGC ((((((........)))))).((((....(((((((........((((..((((((....((((....((........))..))))))))))..)))))))))))...))))........ ( -27.70) >DroYak_CAF1 14403 112 + 1 GCUGUGGCUAAUCAAGAAGCGCCUUAAAGCAUGCUACAUUUU--------UGUGCCAACGUGUACUUCGCAUAUUUCGGCACUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUUC (((..(((............)))......(((((((......--------.(((((((..(((.....)))..))..)))))(((((.....))))).)))))))....)))........ ( -27.60) >consensus GCUUUUGCUAAUCAAGAAGCGCCUUAAAGCAUGCUGCACUUUU_______UGUUGCAACAUGUACUUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUAC ((((((........))))))........(((((.(((.................))).)))))(((((((.((((((.(((((((((.....))))))).)).)))))).)))))))... (-24.91 = -26.95 + 2.04)



| Location | 5,111,607 – 5,111,720 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -22.04 |
| Energy contribution | -23.64 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5111607 113 - 27905053 GUAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAAGUACAUGUUGCAACA-------AAAGCUGCAGCAUGCUUUAAGGCGCUUCUUGAUUAGCAAAAGC (((.(((((((((((...((.((((((.......))))))))..))))).)))))))))((((((((...-------.....))))))))(((....)))(((........)))...... ( -34.20) >DroSec_CAF1 13113 113 - 1 GUAACUUUGCUUUUUCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAGGUACAUGUUGCAACA-------AAAAGUGCAGCAUGCUUUAAGUCGCUUCUUGAUUAGCAAAAGC (((.((((((.(((((..((.((((((.......))))))))..))))).)))))))))((((((((...-------.....))))))))((((((((......)))))..)))...... ( -34.80) >DroSim_CAF1 13664 112 - 1 GUAACUUUGCUUUUUCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAGGUACAUGUUGCAACA-------A-AAGUGCAGCAUGCUUUAAGGCGCCUCUUGAUUAGCAAAAGC (((.((((((.(((((..((.((((((.......))))))))..))))).)))))))))((((((((...-------.-...))))))))(((((((((....))))))..)))...... ( -34.50) >DroEre_CAF1 14440 120 - 1 GCAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAGUGCCGAAAAAUGCGAAAUACAUGUUGGUAAAAAACACAAAAAAUGUAGCAUGCUUUAAGCCGCUUCUUGAUUAGCAAAAGC ....((((((((...(((((((.((((.......)))).((((((...(((.......))).))))))................)))))))....)))).(((........))).)))). ( -25.20) >DroYak_CAF1 14403 112 - 1 GAAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAGUGCCGAAAUAUGCGAAGUACACGUUGGCACA--------AAAAUGUAGCAUGCUUUAAGGCGCUUCUUGAUUAGCCACAGC ........(((((..(((((((.((((.......))))(((((((....(((...)))....))))))).--------......)))))))....)))))(((........)))...... ( -29.10) >consensus GUAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAAGUACAUGUUGCAACA_______AAAAAUGCAGCAUGCUUUAAGGCGCUUCUUGAUUAGCAAAAGC ...((((((((((((...((.((((((.......))))))))..))))).))))))).(((((((((...............))))))))).........(((........)))...... (-22.04 = -23.64 + 1.60)


| Location | 5,111,647 – 5,111,755 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -23.14 |
| Energy contribution | -24.54 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5111647 108 + 27905053 UUU-------UGUUGCAACAUGUACUUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUACGCAUAUGAAAAGUGCUUUGACAACGCCGCAAAAA-C---- (((-------(((.((.....(((((((((.((((((.(((((((((.....))))))).)).)))))).)))))).)))((((.......)))).........)).)))))).-.---- ( -34.30) >DroSec_CAF1 13153 108 + 1 UUU-------UGUUGCAACAUGUACCUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGAAAAAGCAAAGUUACGCAUAUGAAAAGUGCUUUGACAACGCCGCAAAAA-C---- (((-------(((.((.....((((.((((.((((((.(((((((((.....))))))).)).)))))).)))).).)))((((.......)))).........)).)))))).-.---- ( -30.70) >DroSim_CAF1 13704 107 + 1 U-U-------UGUUGCAACAUGUACCUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGAAAAAGCAAAGUUACGCAUAUGAAAAGUGCUUUGACAACGCCGCAAAAA-C---- (-(-------(((.((.....((((.((((.((((((.(((((((((.....))))))).)).)))))).)))).).)))((((.......)))).........)).)))))..-.---- ( -29.80) >DroEre_CAF1 14480 116 + 1 UUUUGUGUUUUUUACCAACAUGUAUUUCGCAUUUUUCGGCACUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUGCGCAUGUGAAAAGUGCUUUGACAUCGCCGCAAAAAAC---- ...((((.....(((......)))...))))(((((((((..(((((.....)))))...(.(((..((((((...(..(....)..)....))))))..)))))))).)))))..---- ( -27.10) >DroYak_CAF1 14443 112 + 1 UU--------UGUGCCAACGUGUACUUCGCAUAUUUCGGCACUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUUCGCAUGUAAAAAGUGCUUUGACACCGGCGCAAAAAAAAAAA ((--------((((((...(((.....))).......((((((((((.....))))....(((((((((.......)))).)))))....))))))........))))))))........ ( -31.20) >consensus UUU_______UGUUGCAACAUGUACUUUGCAUUUUCGGGCAAUGCAGGUAAAUUGCAUUAGCAUGGAAAAGCAAAGUUACGCAUAUGAAAAGUGCUUUGACAACGCCGCAAAAA_C____ ..........(((.((.....(((((((((.((((((.(((((((((.....))))))).)).)))))).)))))).)))((((.......)))).........)).))).......... (-23.14 = -24.54 + 1.40)



| Location | 5,111,647 – 5,111,755 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -21.18 |
| Energy contribution | -22.66 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5111647 108 - 27905053 ----G-UUUUUGCGGCGUUGUCAAAGCACUUUUCAUAUGCGUAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAAGUACAUGUUGCAACA-------AAA ----.-...(((((((((.......(((.........)))(((.(((((((((((...((.((((((.......))))))))..))))).))))))))))))))))))..-------... ( -30.50) >DroSec_CAF1 13153 108 - 1 ----G-UUUUUGCGGCGUUGUCAAAGCACUUUUCAUAUGCGUAACUUUGCUUUUUCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAGGUACAUGUUGCAACA-------AAA ----.-...(((((((((.......(((.........)))(((.((((((.(((((..((.((((((.......))))))))..))))).))))))))))))))))))..-------... ( -31.40) >DroSim_CAF1 13704 107 - 1 ----G-UUUUUGCGGCGUUGUCAAAGCACUUUUCAUAUGCGUAACUUUGCUUUUUCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAGGUACAUGUUGCAACA-------A-A ----.-...(((((((((.......(((.........)))(((.((((((.(((((..((.((((((.......))))))))..))))).))))))))))))))))))..-------.-. ( -31.40) >DroEre_CAF1 14480 116 - 1 ----GUUUUUUGCGGCGAUGUCAAAGCACUUUUCACAUGCGCAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAGUGCCGAAAAAUGCGAAAUACAUGUUGGUAAAAAACACAAAA ----((((((((((((.((((....(((.(((((....((((......((........))...((((.......)))))))).))))).))).....))))))).)))))))))...... ( -27.70) >DroYak_CAF1 14443 112 - 1 UUUUUUUUUUUGCGCCGGUGUCAAAGCACUUUUUACAUGCGAAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAGUGCCGAAAUAUGCGAAGUACACGUUGGCACA--------AA ........((((.((((((((....)))))........(((..(((((((.((((...((...((((.......))))..)).))))...)))))))...))).))).))--------)) ( -28.10) >consensus ____G_UUUUUGCGGCGUUGUCAAAGCACUUUUCAUAUGCGUAACUUUGCUUUUCCAUGCUAAUGCAAUUUACCUGCAUUGCCCGAAAAUGCAAAGUACAUGUUGCAACA_______AAA .........((((((((.(((....(((.........)))...((((((((((((...((.((((((.......))))))))..))))).))))))))))))))))))............ (-21.18 = -22.66 + 1.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:19 2006