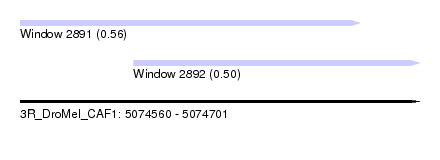
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,074,560 – 5,074,701 |
| Length | 141 |
| Max. P | 0.558882 |

| Location | 5,074,560 – 5,074,680 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -29.27 |
| Energy contribution | -28.50 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
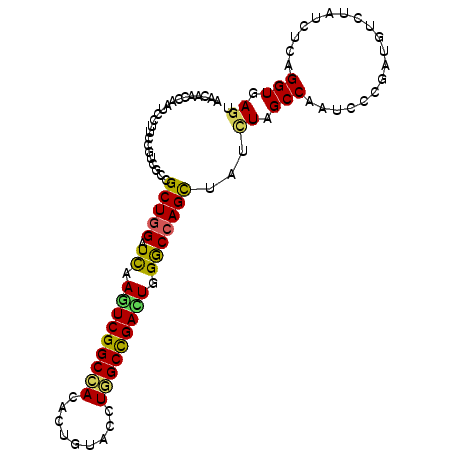
>3R_DroMel_CAF1 5074560 120 + 27905053 CGAUGGCAAGCUGGACUAUGAUAAGCUCUUCGAGUCGCUGAAUAACCAAUCCUUCCGUCGCCGCUGGAUCAAGUCGGCCACACUCUACCUGGCCGACUGGGCCAGCUACCUGGCCAAUCC ((((((.(((...((((.(((........)))))))..((......))...))))))))).....((((..(((((((((.........))))))))).((((((....)))))).)))) ( -41.40) >DroVir_CAF1 895 120 + 1 UGAUGGCAAGUUGGAUUAUGACAAAUUGUUCGAGUCUCUGAACAAUCAAUCAUUCCGUCGUCGCUGGAUCAAAUCGGCCACAUUGUAUUUGGCUGACUGGGCCAGUUAUUUGGCCAAUCC .((((((.....(((..((((...((((((((......))))))))...)))))))))))))...((((....(((((((.........)))))))...((((((....)))))).)))) ( -37.90) >DroGri_CAF1 1845 120 + 1 CGAUGGUAAAGUGGAUUAUGACAAGCUAUUCGAGUCUCUGAACAAUCAAUCCUUCCGUCGUCGCUGGAUCAAGUCGGCCACAUUAUAUUUGGCUGAUUGGACCAGCUAUUUAGCCAAUCC ((((((......(((((..(((...........)))...((....)))))))..))))))..(((((.((.(((((((((.........))))))))).))))))).............. ( -34.30) >DroWil_CAF1 1618 120 + 1 CGAUGGCAAAUUGGACUAUGAUAAGCUCUUUGAAUCUCUGAAUAAUCAAUCGUUCCGUCGCCGCUGGAUUAAAUCGGCCACUUUGUAUUUGGCCGAUUGGGCAAGUUAUCUGGCCAAUCC ....(((....((((..(((((......(((........)))......)))))))))..)))...(((((.(((((((((.........))))))))).(((.((....)).)))))))) ( -30.30) >DroMoj_CAF1 1834 120 + 1 CGAUGGCAAAGUCGAUUACGAUAAACUGUUCGAGUCACUGAAUAAUCAAUCCUUCCGUCGUCGCUGGAUCAAAUCGGCCACUUUGUAUUUGGCUGACUGGGCCAGCUAUAUAGCCAAUCC .((((((..(((.((((.(((........))))))))))((....)).........))))))(((((..((..(((((((.........))))))).))..))))).............. ( -33.90) >DroPer_CAF1 891 120 + 1 CGAUGGAAAGCUGGACUACGACAAGCUCUUCGAAUCGCUGAACAACCAAUCCUUCCGCAGGCGCUGGAUCAAGUCGGCAACGCUUUACCUUGCCGACUGGGCCAGCUAUCUCGCCAAUCC .(((.((((((((........).)))).)))..)))((.(((..........))).)).((((((((..((.((((((((.(.....).))))))))))..)))))......)))..... ( -36.50) >consensus CGAUGGCAAACUGGACUAUGACAAGCUCUUCGAGUCUCUGAACAAUCAAUCCUUCCGUCGUCGCUGGAUCAAAUCGGCCACAUUGUAUUUGGCCGACUGGGCCAGCUAUCUGGCCAAUCC .((((((......((((.(((........)))))))..........................(((((.((.(((((((((.........))))))))).)))))))......))).))). (-29.27 = -28.50 + -0.77)



| Location | 5,074,600 – 5,074,701 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -23.12 |
| Energy contribution | -22.54 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5074600 101 + 27905053 AAUAACCAAUCCUUCCGUCGCCGCUGGAUCAAGUCGGCCACACUCUACCUGGCCGACUGGGCCAGCUACCUGGCCAAUCCCGAGGUGUAUCUCAGGUGAGU .................((((((((((..((.((((((((.........))))))))))..)))))(((((((......)).))))).......))))).. ( -39.70) >DroPse_CAF1 1049 101 + 1 AACAACCAAUCCUUCCGCAGGCGCUGGAUCAAGUCGGCAACGCUUUACCUGGCCGACUGGGCCAGCUAUCUCGCCAAUCCCGAUGUCUAUCUCAGGUGAGU ..........(((.....))).(((((..((.((((((.............))))))))..)))))...((((((...................)))))). ( -29.13) >DroGri_CAF1 1885 101 + 1 AACAAUCAAUCCUUCCGUCGUCGCUGGAUCAAGUCGGCCACAUUAUAUUUGGCUGAUUGGACCAGCUAUUUAGCCAAUCCCGAAAAUUAUCUAAGGUGAGU .....(((..((((........(((((.((.(((((((((.........))))))))).)))))))...............((......)).))))))).. ( -24.70) >DroWil_CAF1 1658 101 + 1 AAUAAUCAAUCGUUCCGUCGCCGCUGGAUUAAAUCGGCCACUUUGUAUUUGGCCGAUUGGGCAAGUUAUCUGGCCAAUCCUGAUGUGUAUUUGAGGUGAGC ...........(((((.(((.(((.(((((.(((((((((.........))))))))).(((.((....)).))))))))....)))....))).).)))) ( -30.20) >DroMoj_CAF1 1874 101 + 1 AAUAAUCAAUCCUUCCGUCGUCGCUGGAUCAAAUCGGCCACUUUGUAUUUGGCUGACUGGGCCAGCUAUAUAGCCAAUCCGGAAAACUAUCUCAGGUGAGC ............(((((....)(((((..((..(((((((.........))))))).))..)))))..............))))......(((....))). ( -23.60) >DroPer_CAF1 931 101 + 1 AACAACCAAUCCUUCCGCAGGCGCUGGAUCAAGUCGGCAACGCUUUACCUUGCCGACUGGGCCAGCUAUCUCGCCAAUCCCGAUGUCUAUCUCAGGUGAGU ..........(((.....))).(((((..((.((((((((.(.....).))))))))))..)))))...((((((...................)))))). ( -31.91) >consensus AACAACCAAUCCUUCCGUCGCCGCUGGAUCAAGUCGGCCACACUGUACCUGGCCGACUGGGCCAGCUAUCUAGCCAAUCCCGAUGUCUAUCUCAGGUGAGU ......................(((((.((.(((((((((.........))))))))).)))))))...((.(((...................))).)). (-23.12 = -22.54 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:39 2006