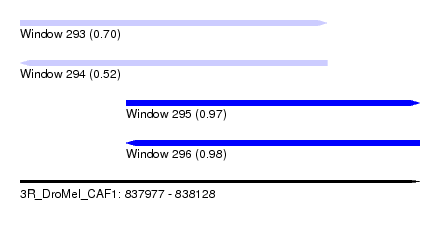
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
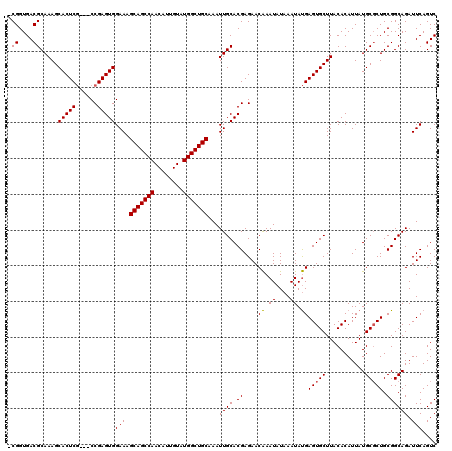
| Location | 837,977 – 838,128 |
| Length | 151 |
| Max. P | 0.976015 |

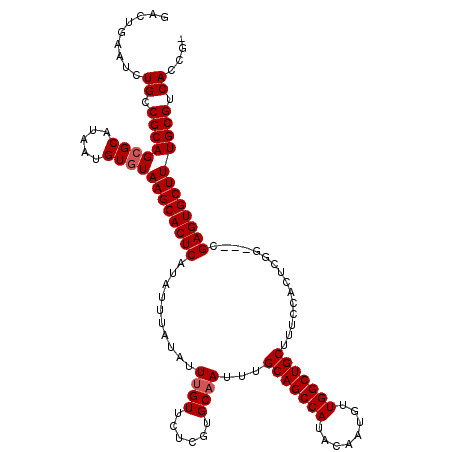
| Location | 837,977 – 838,093 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -33.10 |
| Energy contribution | -33.60 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 837977 116 + 27905053 GACUGAAUCUGCCGCAGCGCAUAAUGUGUAAGCACUCAUAUUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGG---CGAGUGCUUUGCGUCACCG- .........((.((((((((.....))))((((((((..........((((......))))...(((((((........)))))))...........---.)))))))))))).))...- ( -35.40) >DroSec_CAF1 13456 119 + 1 GACUGAAUCUGCCGCAGCGCAUAAUGUGUAAGCACUCAUAUUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGGCGGCGAGUGCUUUGCGUCACCG- .........((.((((((((.....))))(((((((((((....)))..((..(((........(((((((........)))))))........)))..)))))))))))))).))...- ( -35.79) >DroSim_CAF1 12948 119 + 1 GACUGAAUCUGCCGCAGCGCAUAAUGUGUAAGCACUCAUAUUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGGCGGCGAGUGCUUUGCGUCACCG- .........((.((((((((.....))))(((((((((((....)))..((..(((........(((((((........)))))))........)))..)))))))))))))).))...- ( -35.79) >DroEre_CAF1 15097 117 + 1 GACUGAAUCUGCCGCAGAGCAUAAUGUGUAAGCACUCGUAUUUAUAUUCGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGG---UGAGUGCUUUGCGUCACCGA ....((((.(((((.(((((..((((((...((....))...)))))).))))))).)))))))(((((((........))))))).......((((---(((.(((...)))))))))) ( -36.70) >consensus GACUGAAUCUGCCGCAGCGCAUAAUGUGUAAGCACUCAUAUUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGG___CGAGUGCUUUGCGUCACCG_ .........((.((((((((.....))))((((((((..........((((......))))...(((((((........)))))))...............)))))))))))).)).... (-33.10 = -33.60 + 0.50)

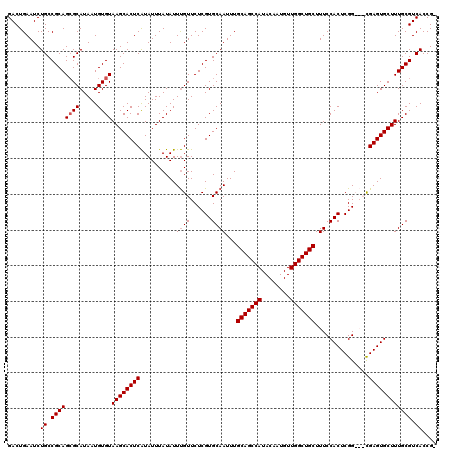
| Location | 837,977 – 838,093 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.35 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 837977 116 - 27905053 -CGGUGACGCAAAGCACUCG---CCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAAUAUGAGUGCUUACACAUUAUGCGCUGCGGCAGAUUCAGUC -((....))..........(---(((((((.(..(((((((........))))))).....((((....................))))..........).)))).)))).......... ( -32.15) >DroSec_CAF1 13456 119 - 1 -CGGUGACGCAAAGCACUCGCCGCCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAAUAUGAGUGCUUACACAUUAUGCGCUGCGGCAGAUUCAGUC -.(((((.((...))..)))))((((((((.(..(((((((........))))))).....((((....................))))..........).)))).)))).......... ( -35.85) >DroSim_CAF1 12948 119 - 1 -CGGUGACGCAAAGCACUCGCCGCCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAAUAUGAGUGCUUACACAUUAUGCGCUGCGGCAGAUUCAGUC -.(((((.((...))..)))))((((((((.(..(((((((........))))))).....((((....................))))..........).)))).)))).......... ( -35.85) >DroEre_CAF1 15097 117 - 1 UCGGUGACGCAAAGCACUCA---CCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACGAAUAUAAAUACGAGUGCUUACACAUUAUGCUCUGCGGCAGAUUCAGUC (((((((.((...))..)))---))))(((....(((((((........))))))).....((((....................))))...)))....((((....))))......... ( -32.75) >consensus _CGGUGACGCAAAGCACUCG___CCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAAUAUGAGUGCUUACACAUUAUGCGCUGCGGCAGAUUCAGUC .((....)).....(((((......)))))(((.(((((((........)))))))...((((.((....((.((....)).))(((((...........))))).)))))).))).... (-31.48 = -31.35 + -0.13)

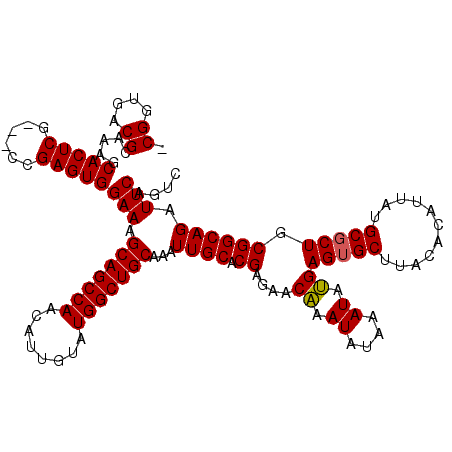
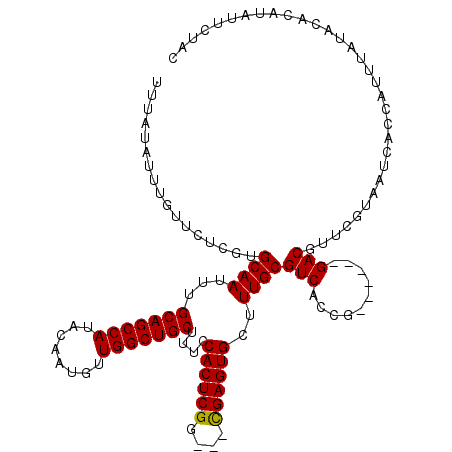
| Location | 838,017 – 838,128 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -25.14 |
| Energy contribution | -24.95 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 838017 111 + 27905053 UUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGG---CGAGUGCUUUGCGUCACCG------GACGUUUGUAAUCACCAUUUUUACAUAUAUUCUAC ...((((.(((....(((.((...(((((((........))))))).)).)))..((---.((.(((...(((((....------)))))..))).)).))......))).))))..... ( -30.40) >DroSec_CAF1 13496 114 + 1 UUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGGCGGCGAGUGCUUUGCGUCACCG------GACGUUCGUAAUCACCAUUUAUACACAUAUUCUAC ...((((.(((....(((.((...(((((((........))))))).)).))).....((.((.(((...(((((....------)))))..))).)).))......))).))))..... ( -29.50) >DroSim_CAF1 12988 114 + 1 UUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGGCGGCGAGUGCUUUGCGUCACCG------GACGUUCGUAAUCACCAUUUAUACACAUAUUCUAC ...((((.(((....(((.((...(((((((........))))))).)).))).....((.((.(((...(((((....------)))))..))).)).))......))).))))..... ( -29.50) >DroEre_CAF1 15137 113 + 1 UUUAUAUUCGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGG---UGAGUGCUUUGCGUCACCGAUUAUGGACGUUCAUAAUCGCCAUUU----AAAUAUUCCAC .((((...(((((..(((.((...(((((((........))))))).)).)))((((---(((.(((...))))))))))....)))))...)))).........----........... ( -32.80) >consensus UUUAUAUUUGUUCUCGUGCAAUUUGCAGCCAUACAAUGUUGGCUGCUUUCCACUCGG___CGAGUGCUUUGCGUCACCG______GACGUUCGUAAUCACCAUUUAUACACAUAUUCUAC .................((((...(((((((........)))))))....((((((....))))))..))))(((..........)))................................ (-25.14 = -24.95 + -0.19)

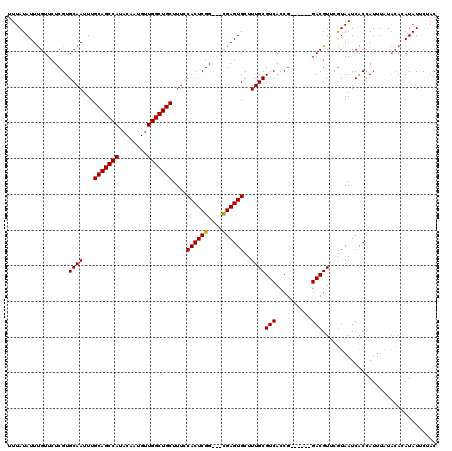
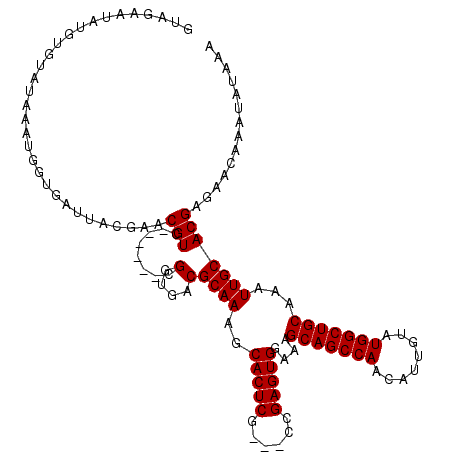
| Location | 838,017 – 838,128 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 838017 111 - 27905053 GUAGAAUAUAUGUAAAAAUGGUGAUUACAAACGUC------CGGUGACGCAAAGCACUCG---CCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAA .....((((.(((.......(..(((.....((..------((((((.((...))..)))---)))..))....(((((((........))))))).)))..)......))).))))... ( -29.02) >DroSec_CAF1 13496 114 - 1 GUAGAAUAUGUGUAUAAAUGGUGAUUACGAACGUC------CGGUGACGCAAAGCACUCGCCGCCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAA .....((((.(((......((((........)).)------)(....)((((..((((((....))))))....(((((((........)))))))...))))......))).))))... ( -29.70) >DroSim_CAF1 12988 114 - 1 GUAGAAUAUGUGUAUAAAUGGUGAUUACGAACGUC------CGGUGACGCAAAGCACUCGCCGCCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAA .....((((.(((......((((........)).)------)(....)((((..((((((....))))))....(((((((........)))))))...))))......))).))))... ( -29.70) >DroEre_CAF1 15137 113 - 1 GUGGAAUAUUU----AAAUGGCGAUUAUGAACGUCCAUAAUCGGUGACGCAAAGCACUCA---CCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACGAAUAUAAA .....((((((----...((((((((.......(((((..(((((((.((...))..)))---)))))))))..(((((((........))))))).)))))).)).....))))))... ( -34.80) >consensus GUAGAAUAUGUGUAUAAAUGGUGAUUACGAACGUC______CGGUGACGCAAAGCACUCG___CCGAGUGGAAAGCAGCCAACAUUGUAUGGCUGCAAAUUGCACGAGAACAAAUAUAAA ...............................(((........(....)((((..(((((......)))))....(((((((........)))))))...))))))).............. (-27.48 = -27.47 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:01 2006