
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,070,846 – 5,070,954 |
| Length | 108 |
| Max. P | 0.731176 |

| Location | 5,070,846 – 5,070,954 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.65 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5070846 108 + 27905053 CAAUACGGUGAGAAAUGGUCGUGUGGAUACGCGAUCAUAGAGACUGUGCUUCACAUACUCCCGCAUUAUGUAGGCUGCCUUGUCGAUAAGCUAAAAAUGAUGA-UUAAU .....(((.(((..((((((((((....))))))))))......((((...))))..))))))((((((...((((............))))....)))))).-..... ( -27.80) >DroVir_CAF1 4094 108 + 1 GAGCACAGUAAGAAAAGGUCGAGUGGACACGCGAUCGUACAGACUGUGCUUGACAUACUCUCGCAUUAUGUAGGCUGCCUUGUCGAUGAGCUGUUGUGGA-AUGUUAAA (((((((((.......(((((.((....)).)))))......)))))))))(((((..((.((((....((.(((......))).....))...))))))-)))))... ( -30.22) >DroGri_CAF1 4121 105 + 1 CAGCACAGUUAAAAAUGGUCGCGUGGACACGCGAUCAUAAAGACUAUGCUUCACAUACUCGCGCAUUAUGUAGGCCGCCUUAUCAAUGAGCUGGAAAGGA----UUAAA .((((.((((....((((((((((....))))))))))...)))).)))).......((.((.(((((((.(((...)))))).)))).)).))......----..... ( -28.80) >DroEre_CAF1 4284 108 + 1 CAACACGGUGAGAAAUGGGCGUGUGGAUACGCGGUCAUAAAGACUGUGUUUCACAUACUCCCGCAUUAUGUAGGCUGCUUUGUCUAUUAGCUGAAAAGUAUGA-AUAAU ..(((..(((.....((((.(((((((.((((((((.....)))))))).)).))))).)))))))..)))((((......))))....(((....)))....-..... ( -34.80) >DroYak_CAF1 4275 108 + 1 CAGUACGGUGAGAAAUGGGCGUGUGGACACGCGGUCAUAAAGACUGUGUUUCACAUACUCCCGCAUUAUGUAGGCUGCCUUGUCGAUAAGCUGAAAAAUAUGA-UUAAU ((((.(((..((....(((.(((((((.((((((((.....)))))))).)).))))).)))(((..........)))))..)))....))))..........-..... ( -34.80) >DroMoj_CAF1 4158 102 + 1 CAGCACAGUGAGAAAAGGUCGCGUGGAAACGCGGUCGUAUAGACUAUGCUUAACGUACUGCCGCAUUAUGUAGGCUGCUUUGUCAAUGAGCUGGUAUGAA-AA------ .((((.(((.......(..(((((....)))))..)......))).))))...(((((((((((.....)).))).((((.......)))).))))))..-..------ ( -31.92) >consensus CAGCACAGUGAGAAAUGGUCGUGUGGACACGCGAUCAUAAAGACUGUGCUUCACAUACUCCCGCAUUAUGUAGGCUGCCUUGUCGAUGAGCUGAAAAGGA_GA_UUAAU .((((((((.....((((((((((....))))))))))....))))))))............((.((((...(((......))).)))))).................. (-19.90 = -20.65 + 0.75)
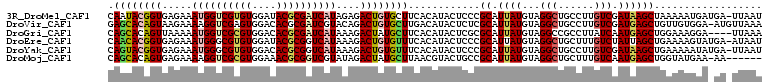
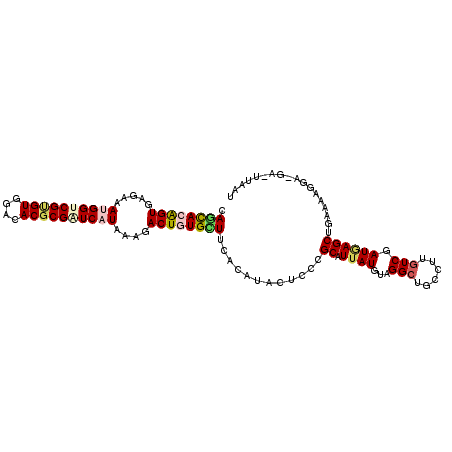

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:33 2006