
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,059,324 – 5,059,432 |
| Length | 108 |
| Max. P | 0.707585 |

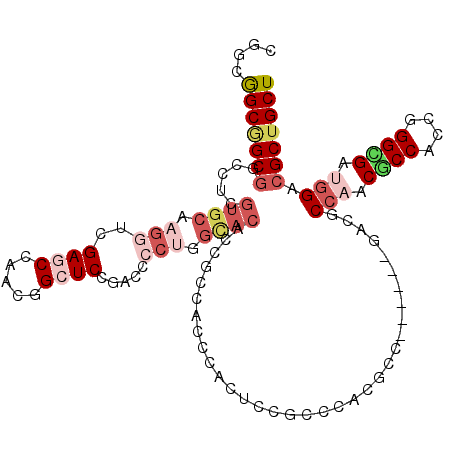
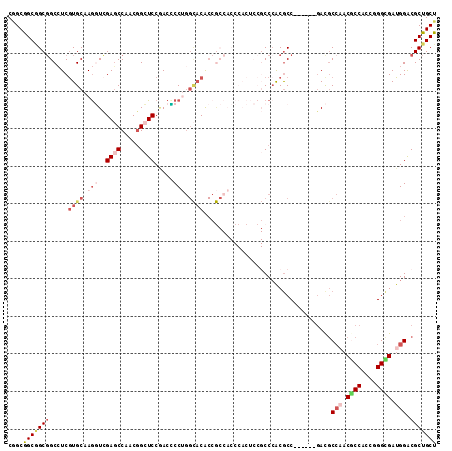
| Location | 5,059,324 – 5,059,432 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -25.27 |
| Energy contribution | -27.38 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5059324 108 + 27905053 CGGCGGCGGCGGCCUCGUGCAAGGUCGAGCCUACGGCUCCGACCCCUGGCACACCGCCACCCACUCCGCCCACGCC------GACGCCAACCCCACCAGGCGAUGGACGCUGCU .(((((((((((....((((..(((((((((...)))).)))))....)))).))))).........(.(((((((------................)))).))).))))))) ( -48.19) >DroSec_CAF1 165684 108 + 1 CGGCGGCGGCGGCCUCGUGCAAGGUCGAGCCUACGGCUCCGACCCCUGGCACACCGCCACCCACUCCGCCCACGCC------GACGCCAACGCCACCAGGCGAUGGACGCUGCU .(((((.((.(((...((((..(((((((((...)))).)))))....))))...))).))....)))))...((.------(.(((((.((((....)))).))).))).)). ( -51.80) >DroSim_CAF1 171513 108 + 1 CGGCGGCGGCGGCCUCGUGCAAGGUCGAGCCUACGGCUCCGACUCCUGGCACACCGCCACCCACUCCGCCCACGCC------GACGCCAACGCCACCGGGCGAUGGACGCUGCU .(((((.((.(((...((((..(((((((((...)))).)))))....))))...))).))....)))))...((.------(.(((((.((((....)))).))).))).)). ( -49.80) >DroYak_CAF1 172476 114 + 1 CCGCGGCAGCGGCCUCGUGCAAGGUCGAGCCAACGGCUCCCACCCCUGGCACACCGCCACCCACUCCGCCCACGCCCACUCCGACGCCAACGCCACCGGGCGAUGGACGCUGCU ....(((((((((...((((.(((..(((((...))))).....))).))))...))).........(.((((((((....((.......)).....))))).))).))))))) ( -43.30) >DroMoj_CAF1 206943 87 + 1 CAGCAGCCGCGGCGUCGUGCAAGUUGGAGCCAACGGGUCCGACGCCGA---CG---------------CCAGCGCC------GAUGC---CGCCGCCGGGUGAUGGGCGCUGCU .((((((.(((((((((.((..((((((.((...)).)))))))))))---))---------------)).))(((------.((..---((((....)))))).))))))))) ( -49.50) >DroAna_CAF1 160296 96 + 1 CGGCGGCAGCUGCAUCAUGCAAAUUGGAUCCAACUGCUCCAACACCCGGUACACCA------------CCCACACC------CACACCCACUCCUCCCGGAGAUGGGCGCUGCU ....(((((((((.....)))..(((((.(.....).))))).....((....)).------------........------....((((((((....)))).)))).)))))) ( -27.40) >consensus CGGCGGCGGCGGCCUCGUGCAAGGUCGAGCCAACGGCUCCGACCCCUGGCACACCGCCACCCACUCCGCCCACGCC______GACGCCAACGCCACCGGGCGAUGGACGCUGCU ....(((((((.....((((.(((..((((.....)))).....))).))))..................................(((.((((....)))).))).))))))) (-25.27 = -27.38 + 2.11)

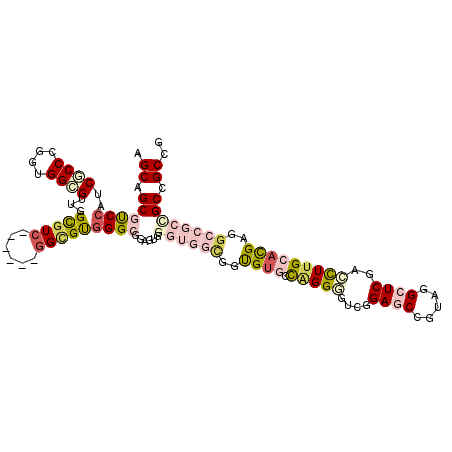
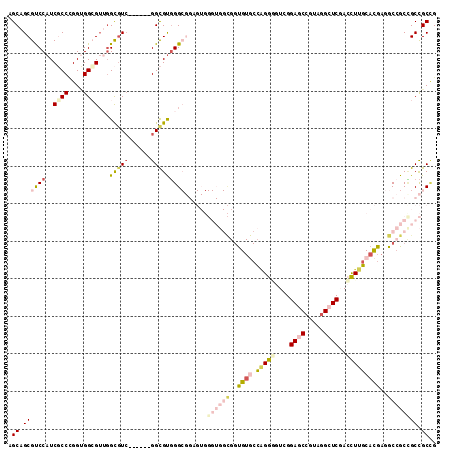
| Location | 5,059,324 – 5,059,432 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -55.38 |
| Consensus MFE | -30.36 |
| Energy contribution | -32.90 |
| Covariance contribution | 2.54 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5059324 108 - 27905053 AGCAGCGUCCAUCGCCUGGUGGGGUUGGCGUC------GGCGUGGGCGGAGUGGGUGGCGGUGUGCCAGGGGUCGGAGCCGUAGGCUCGACCUUGCACGAGGCCGCCGCCGCCG .((..((((((.((((.(..(.......)..)------))))))))))..)).((((((((((.(((.(((((((.((((...)))))))))))......))))))))))))). ( -59.50) >DroSec_CAF1 165684 108 - 1 AGCAGCGUCCAUCGCCUGGUGGCGUUGGCGUC------GGCGUGGGCGGAGUGGGUGGCGGUGUGCCAGGGGUCGGAGCCGUAGGCUCGACCUUGCACGAGGCCGCCGCCGCCG .((.(((.(((.((((....)))).)))))).------.))...(((((....((((((..(((((..(((((((.((((...))))))))))))))))..)))))).))))). ( -63.00) >DroSim_CAF1 171513 108 - 1 AGCAGCGUCCAUCGCCCGGUGGCGUUGGCGUC------GGCGUGGGCGGAGUGGGUGGCGGUGUGCCAGGAGUCGGAGCCGUAGGCUCGACCUUGCACGAGGCCGCCGCCGCCG .((.(((.(((.((((....)))).)))))).------.))...(((((....((((((..(((((.(((.((((.((((...))))))))))))))))..)))))).))))). ( -59.70) >DroYak_CAF1 172476 114 - 1 AGCAGCGUCCAUCGCCCGGUGGCGUUGGCGUCGGAGUGGGCGUGGGCGGAGUGGGUGGCGGUGUGCCAGGGGUGGGAGCCGUUGGCUCGACCUUGCACGAGGCCGCUGCCGCGG .((..((((((.((((((.(((((....)))))...)))))))))))).....((..(((((((((.((((....(((((...)))))..))))))))...)))))..)))).. ( -61.40) >DroMoj_CAF1 206943 87 - 1 AGCAGCGCCCAUCACCCGGCGGCG---GCAUC------GGCGCUGG---------------CG---UCGGCGUCGGACCCGUUGGCUCCAACUUGCACGACGCCGCGGCUGCUG ((((((.........((((((.((---....)------).))))))---------------((---.((((((((..(..((((....))))..)..)))))))))))))))). ( -47.70) >DroAna_CAF1 160296 96 - 1 AGCAGCGCCCAUCUCCGGGAGGAGUGGGUGUG------GGUGUGGG------------UGGUGUACCGGGUGUUGGAGCAGUUGGAUCCAAUUUGCAUGAUGCAGCUGCCGCCG .((.(((((((.((((....))))))))))).------.))...((------------((((...((((.(((....))).)))).......((((.....))))..)))))). ( -41.00) >consensus AGCAGCGUCCAUCGCCCGGUGGCGUUGGCGUC______GGCGUGGGCGGAGUGGGUGGCGGUGUGCCAGGGGUCGGAGCCGUAGGCUCGACCUUGCACGAGGCCGCCGCCGCCG .((.((((((..((((....))))...(((((......)))))))))......((((((..((((.(((((....((((.....))))..)))))))))..)))))))).)).. (-30.36 = -32.90 + 2.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:30 2006