
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,053,303 – 5,053,397 |
| Length | 94 |
| Max. P | 0.997033 |

| Location | 5,053,303 – 5,053,397 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5053303 94 + 27905053 AUGCUCACUAUCCAAUCGCCCGUCUUAACCUCCAAGUAAUUAAAUUGCUCGCGACAGUUGCUGGCAAUUAAAAUCGUUUGGAGAGCCAAGUCAA .....................(.(((..((((((((..(((.(((((((.((((...)))).)))))))..)))..))))))).)..))).).. ( -22.20) >DroSec_CAF1 159534 94 + 1 AUGCUCACUAUCCAAUCGCCCGUCUUAACCUCCAAGUAAUUAAAUUGCUCGCGACAGUUGCUGGCAAUUAAAAUCGUUUGGAGAGCCAAGUCAA .....................(.(((..((((((((..(((.(((((((.((((...)))).)))))))..)))..))))))).)..))).).. ( -22.20) >DroSim_CAF1 165249 94 + 1 AUGCUCACUAUCCAAUCGCCCGUCUUAACCUCCAAGUAAUUAAAUUGCUCGCGACAGUUGCUGGCAAUUAAAAUCGUUUGGAGAGCCAAGUCAA .....................(.(((..((((((((..(((.(((((((.((((...)))).)))))))..)))..))))))).)..))).).. ( -22.20) >DroEre_CAF1 162011 89 + 1 AUGCUCACCA-----UCGCCCGUCUUAACCUCCAAGUAAUUAAAUUGCUCGCGACAGUUGCUGGCAAUUAAAAUAGUUUGGAGAGCCAAGUCAA ..........-----......(.(((..((((((((..(((.(((((((.((((...)))).)))))))..)))..))))))).)..))).).. ( -20.80) >DroYak_CAF1 166101 90 + 1 AUGCUCAUGAU----UCGCCCGUCUUAACCUCCAAGUAAUUAAAUUGCUCGCGACAGUUGCUGGCAAUUAAAAUCGUUUGGAGAGCCAAGUCAA ..((((..(((----......)))......((((((..(((.(((((((.((((...)))).)))))))..)))..))))))))))........ ( -22.90) >consensus AUGCUCACUAUCCAAUCGCCCGUCUUAACCUCCAAGUAAUUAAAUUGCUCGCGACAGUUGCUGGCAAUUAAAAUCGUUUGGAGAGCCAAGUCAA .....................(.(((..((((((((..(((.(((((((.((((...)))).)))))))..)))..))))))).)..))).).. (-21.92 = -21.92 + -0.00)

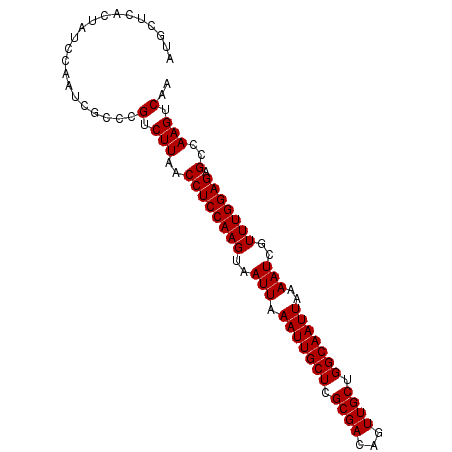
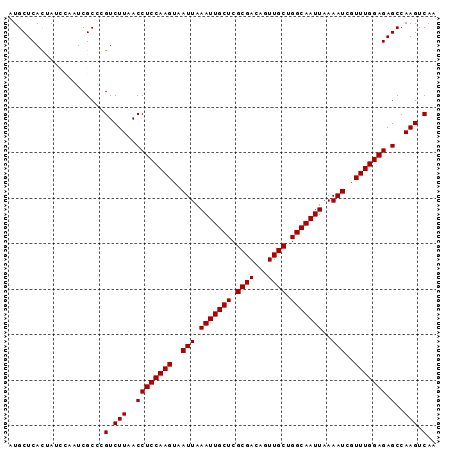
| Location | 5,053,303 – 5,053,397 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5053303 94 - 27905053 UUGACUUGGCUCUCCAAACGAUUUUAAUUGCCAGCAACUGUCGCGAGCAAUUUAAUUACUUGGAGGUUAAGACGGGCGAUUGGAUAGUGAGCAU ....((((((.((((((..((((..((((((..((.......))..)))))).))))..))))))))))))....((.............)).. ( -24.62) >DroSec_CAF1 159534 94 - 1 UUGACUUGGCUCUCCAAACGAUUUUAAUUGCCAGCAACUGUCGCGAGCAAUUUAAUUACUUGGAGGUUAAGACGGGCGAUUGGAUAGUGAGCAU ....((((((.((((((..((((..((((((..((.......))..)))))).))))..))))))))))))....((.............)).. ( -24.62) >DroSim_CAF1 165249 94 - 1 UUGACUUGGCUCUCCAAACGAUUUUAAUUGCCAGCAACUGUCGCGAGCAAUUUAAUUACUUGGAGGUUAAGACGGGCGAUUGGAUAGUGAGCAU ....((((((.((((((..((((..((((((..((.......))..)))))).))))..))))))))))))....((.............)).. ( -24.62) >DroEre_CAF1 162011 89 - 1 UUGACUUGGCUCUCCAAACUAUUUUAAUUGCCAGCAACUGUCGCGAGCAAUUUAAUUACUUGGAGGUUAAGACGGGCGA-----UGGUGAGCAU ....((((((.((((((...(((..((((((..((.......))..)))))).)))...))))))))))))....((..-----......)).. ( -24.40) >DroYak_CAF1 166101 90 - 1 UUGACUUGGCUCUCCAAACGAUUUUAAUUGCCAGCAACUGUCGCGAGCAAUUUAAUUACUUGGAGGUUAAGACGGGCGA----AUCAUGAGCAU ....((((((.((((((..((((..((((((..((.......))..)))))).))))..))))))))))))....((..----.......)).. ( -25.00) >consensus UUGACUUGGCUCUCCAAACGAUUUUAAUUGCCAGCAACUGUCGCGAGCAAUUUAAUUACUUGGAGGUUAAGACGGGCGAUUGGAUAGUGAGCAU ....((((((.((((((..((((..((((((..((.......))..)))))).))))..))))))))))))....((.............)).. (-24.00 = -24.20 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:24 2006