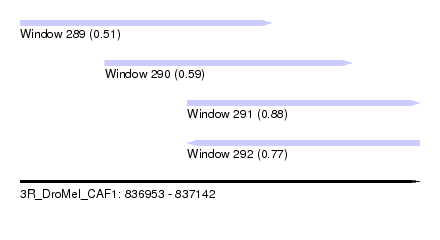
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 836,953 – 837,142 |
| Length | 189 |
| Max. P | 0.882768 |

| Location | 836,953 – 837,072 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -22.65 |
| Energy contribution | -24.52 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 836953 119 + 27905053 CAUCUCUGCGGUUGCCAAUCAAUAAUUUGCCUGGUGAAUGCCUCCGCU-GGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACU .......((((..((((....((((((.(((.((((........))))-)))...))))))....))))..))))...((((...(((((((........)))))))..))))....... ( -31.90) >DroSec_CAF1 12341 119 + 1 CAUCUCUGCGGUUGCCAAUCAAUAAUUUGCCUGGUGAAUGCCUCCGCU-GGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACU .......((((..((((....((((((.(((.((((........))))-)))...))))))....))))..))))...((((...(((((((........)))))))..))))....... ( -31.90) >DroSim_CAF1 11830 119 + 1 CAUCUCUGCGGUUGCCAAUCAAUAAUUUGCCUGGUGAAUGCCUCCGCU-GGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACU .......((((..((((....((((((.(((.((((........))))-)))...))))))....))))..))))...((((...(((((((........)))))))..))))....... ( -31.90) >DroEre_CAF1 14052 119 + 1 CAUCUCUGUGGUUGCCAAUCAAUAAUUUGCCUGGUGAAUGCCUCCGCU-GGCUGUAAUUGUGUUCUGGCCUCUGCCUUUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACU .......(..(..((((....((((((.(((.((((........))))-)))...))))))....))))..)..).((((((...(((((((........)))))))..))))))..... ( -30.60) >DroYak_CAF1 12984 120 + 1 CAUCUCUGCGGUUGCCAAUCAAUAAUUUGCCUGGUGAAUGCCUCCGCUCGGUUGUAAUUGUGAUNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ((.((..(((((..(((..(((....)))..)))..)......))))..)).)).................................................................. ( -12.80) >consensus CAUCUCUGCGGUUGCCAAUCAAUAAUUUGCCUGGUGAAUGCCUCCGCU_GGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACU .......((((..((((....((((((.(((.((((........)))).)))...))))))....))))..))))...((((...(((((((........)))))))..))))....... (-22.65 = -24.52 + 1.87)

| Location | 836,993 – 837,110 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.19 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -29.31 |
| Energy contribution | -29.25 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 836993 117 + 27905053 CCUCCGCUGGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCUUCCACUGCUCUGCAGAGAGAGAAA-- .(((.((.((((..(((...)))..))))...))((((((....(((((((........))))))).(((.((((((((.......))))))))))).......)))))).)))...-- ( -35.00) >DroSec_CAF1 12381 117 + 1 CCUCCGCUGGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGCUCUGCAUAGAGAGAAA-- ....((.(((.(((((((......((((...(((...((((...(((((((........)))))))..)))).)))...))))...))))))).))).))((((......))))...-- ( -28.70) >DroSim_CAF1 11870 117 + 1 CCUCCGCUGGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGCUUUGCAGAGAGAGAAA-- .((((..(((.(((((((......((((...(((...((((...(((((((........)))))))..)))).)))...))))...))))))).)))((((...)))).).)))...-- ( -31.40) >DroEre_CAF1 14092 119 + 1 CCUCCGCUGGCUGUAAUUGUGUUCUGGCCUCUGCCUUUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGUCCUACAGAGAGAGAAACU .((((..(((.(((((((......((((...(((..(((((...(((((((........)))))))..))))))))...))))...))))))).)))(((.....))).).)))..... ( -31.00) >consensus CCUCCGCUGGCUGUAAUUGUGUUCUGGCCUCUGCCUCUGUUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGCUCUGCAGAGAGAGAAA__ .(((.((.((((..(((...)))..))))...))((((((....(((((((........))))))).(((...((((((.......))))))..))).......)))))).)))..... (-29.31 = -29.25 + -0.06)

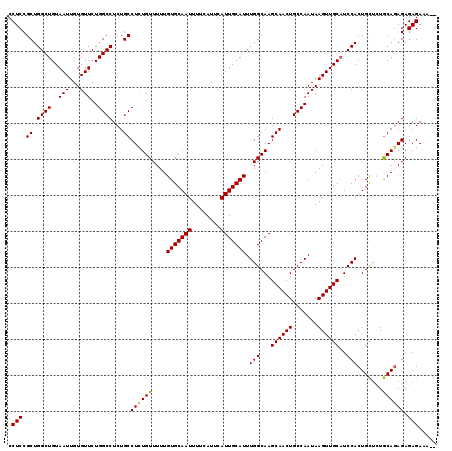
| Location | 837,032 – 837,142 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 837032 110 + 27905053 UUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCUUCCACUGCUCUGCAGAGAGAGAAA-------UGCG---GGUGUACUUAAGUAUCUCAAUAUAAUUU .......(((.(((((............(((.((((((((.......)))))))))))((((...))))...))))).-------)))(---((.((((....))))))).......... ( -29.20) >DroSec_CAF1 12420 110 + 1 UUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGCUCUGCAUAGAGAGAAA-------UGCG---GAUGUACUUAAGUAUCUCAAUAUAAUUU .....(((((((........)))))))((((((......))))))(((((.((((((.(..((((......))))...-------.).)---)))))))))).................. ( -29.30) >DroSim_CAF1 11909 110 + 1 UUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGCUUUGCAGAGAGAGAAA-------UGCG---GAUGUACUUAAGUAUCUCAAUAUAAUUA .....(((((((........)))))))((((((......))))))(((((.((((((.((((...))))....(....-------)..)---)))))))))).................. ( -27.20) >DroEre_CAF1 14131 118 + 1 UUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGUCCUACAGAGAGAGAAACUGAAAAUGUGAAGCAUUUAUUUAAACUUCUGAAUAU--UUA .....((((.((((((((((..((((.((((((......)))))).....))))(((.(((.....))).).)))))..))))))).....)))).((((((......)))))).--... ( -22.60) >consensus UUUUUGUGCAAUUUUCAUUCAUUGCAUUUGGCAAGCAACUGCCAAUAAGUUGCAUCCACUGCUCUGCAGAGAGAGAAA_______UGCG___GAUGUACUUAAGUAUCUCAAUAUAAUUA .....(((((((........)))))))...(((.((((((.......)))))).(((.((((...)))).).))...........)))....((.((((....)))).)).......... (-20.52 = -20.02 + -0.50)

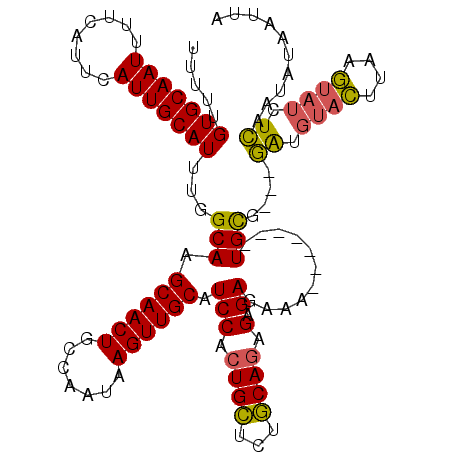
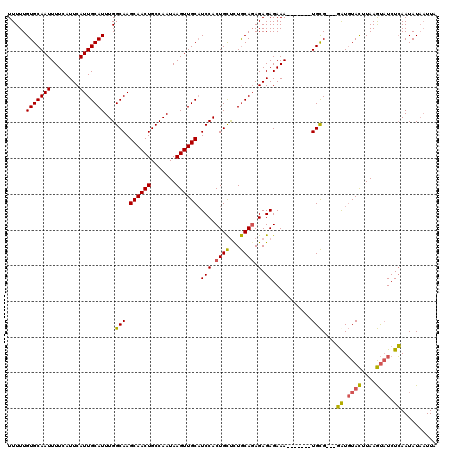
| Location | 837,032 – 837,142 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 837032 110 - 27905053 AAAUUAUAUUGAGAUACUUAAGUACACC---CGCA-------UUUCUCUCUCUGCAGAGCAGUGGAAGCAACUUAUUGGCAGUUGCUUGCCAAAUGCAAUGAAUGAAAAUUGCACAAAAA ........(((...(((....)))....---.(((-------((((..((..((((......(((((((((((.......)))))))).)))..))))..))..))))..))).)))... ( -24.80) >DroSec_CAF1 12420 110 - 1 AAAUUAUAUUGAGAUACUUAAGUACAUC---CGCA-------UUUCUCUCUAUGCAGAGCAGUGGAUGCAACUUAUUGGCAGUUGCUUGCCAAAUGCAAUGAAUGAAAAUUGCACAAAAA ........(((.......(((((.((((---(((.-------...((((......))))..)))))))..)))))(((((((....))))))).((((((........)))))))))... ( -28.60) >DroSim_CAF1 11909 110 - 1 UAAUUAUAUUGAGAUACUUAAGUACAUC---CGCA-------UUUCUCUCUCUGCAAAGCAGUGGAUGCAACUUAUUGGCAGUUGCUUGCCAAAUGCAAUGAAUGAAAAUUGCACAAAAA ........(((.......(((((.((((---(((.-------..........(((...))))))))))..)))))(((((((....))))))).((((((........)))))))))... ( -24.60) >DroEre_CAF1 14131 118 - 1 UAA--AUAUUCAGAAGUUUAAAUAAAUGCUUCACAUUUUCAGUUUCUCUCUCUGUAGGACAGUGGAUGCAACUUAUUGGCAGUUGCUUGCCAAAUGCAAUGAAUGAAAAUUGCACAAAAA ...--.......(((((..........)))))........((((.(..((.(((.....))).))..).))))..(((((((....))))))).((((((........))))))...... ( -22.80) >consensus AAAUUAUAUUGAGAUACUUAAGUACAUC___CGCA_______UUUCUCUCUCUGCAGAGCAGUGGAUGCAACUUAUUGGCAGUUGCUUGCCAAAUGCAAUGAAUGAAAAUUGCACAAAAA ................................(((.......((((..((..((((......(((..((((((.......))))))...)))..))))..))..))))..)))....... (-18.41 = -18.47 + 0.06)


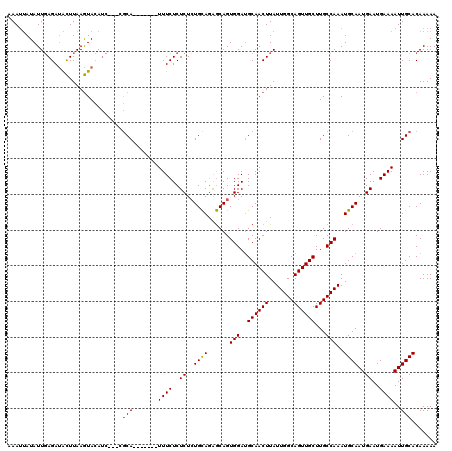
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:54 2006