
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,048,127 – 5,048,275 |
| Length | 148 |
| Max. P | 0.999043 |

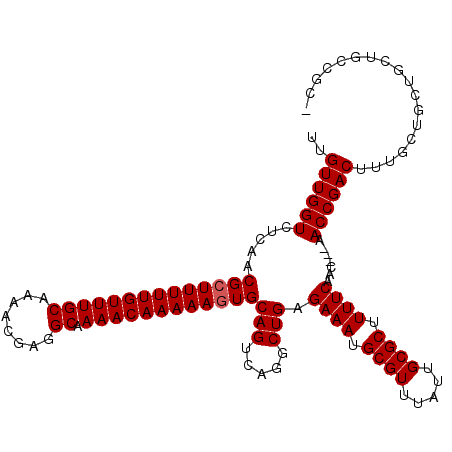
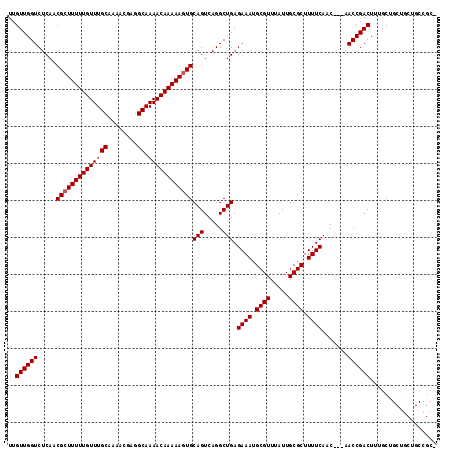
| Location | 5,048,127 – 5,048,239 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -30.08 |
| Energy contribution | -30.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5048127 112 + 27905053 UUGUUGGUCUCAACGCUUUUUGUUUGCAAAACGAGGCAAAACAAAAAGUGCAGUCAGGCUGAGAAAUGCGUUUAUUGCGCUUUUCAAC---AACCGACUUUGCUGCUGCUGCCGC- ..((((((.....((((((((((((((........)).))))))))))))(((.....))).((((.((((.....)))).))))...---.))))))...((.((....)).))- ( -34.00) >DroSec_CAF1 154400 112 + 1 UUGUUGGUCUCAACGCUUUUUGUUUGCAAAACGAGGCAAAACAAAAAGUGCAGUCAGGCUGAGAAAUGCGUUUAUUGCGCUUUUCAAC---AACCGACUUUGCUGCUGCUGCCGC- ..((((((.....((((((((((((((........)).))))))))))))(((.....))).((((.((((.....)))).))))...---.))))))...((.((....)).))- ( -34.00) >DroSim_CAF1 160190 113 + 1 UUGUUGGUCUCAACGGUUUUUGUUUGCAAAACGAGGCAAAACAAAAAGUGCAGUCAGGCUGAGAAAUGCGUUUAUUGCGCUUUUCAAC---AACCGACUUUGCUGCUGCUGCCGCU ..((((((.....((.(((((((((((........)).))))))))).))(((.....))).((((.((((.....)))).))))...---.))))))...((.((....)).)). ( -31.00) >DroEre_CAF1 156786 103 + 1 UUGUUGGUCUCAACGCUUUUUGUUUGCAAAACGAGGCAAAACAAAAAGUGCAGUCAGGCUGAGAAAUGCGUUUAUUGCGCUUUUCAACAACAACCGACUUUGC------------- ..((((((.....((((((((((((((........)).))))))))))))(((.....))).((((.((((.....)))).)))).......)))))).....------------- ( -31.20) >DroYak_CAF1 160932 100 + 1 UUGUUGGUCUCAACGCUUUUUGUUUGCAAAACGAGGCAAAACAAAAAGUGCAGUCAGGCUGAGAAAUGCGUUUAUUGCGCUUUUCAAC---AACCGACUUUGC------------- ..((((((.....((((((((((((((........)).))))))))))))(((.....))).((((.((((.....)))).))))...---.)))))).....------------- ( -31.20) >consensus UUGUUGGUCUCAACGCUUUUUGUUUGCAAAACGAGGCAAAACAAAAAGUGCAGUCAGGCUGAGAAAUGCGUUUAUUGCGCUUUUCAAC___AACCGACUUUGCUGCUGCUGCCGC_ ..((((((.....((((((((((((((........)).))))))))))))(((.....))).((((.((((.....)))).)))).......)))))).................. (-30.08 = -30.28 + 0.20)

| Location | 5,048,127 – 5,048,239 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5048127 112 - 27905053 -GCGGCAGCAGCAGCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUUUUGCCUCGUUUUGCAAACAAAAAGCGUUGAGACCAACAA -..((...((((.(((..(((((((---(..((((.(((.......))).)))).))))).)))))).((((((((((.((........)))))))))))).))))...))..... ( -35.30) >DroSec_CAF1 154400 112 - 1 -GCGGCAGCAGCAGCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUUUUGCCUCGUUUUGCAAACAAAAAGCGUUGAGACCAACAA -..((...((((.(((..(((((((---(..((((.(((.......))).)))).))))).)))))).((((((((((.((........)))))))))))).))))...))..... ( -35.30) >DroSim_CAF1 160190 113 - 1 AGCGGCAGCAGCAGCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUUUUGCCUCGUUUUGCAAACAAAAACCGUUGAGACCAACAA ...((...((((.(((..(((((((---(..((((.(((.......))).)))).))))).))))))..(((((((((.((........)))))))))))..))))...))..... ( -32.40) >DroEre_CAF1 156786 103 - 1 -------------GCAAAGUCGGUUGUUGUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUUUUGCCUCGUUUUGCAAACAAAAAGCGUUGAGACCAACAA -------------(((..(((((..((((..((((.(((.......))).)))).)))))))))))).((((((((((.((........)))))))))))).((((....)))).. ( -30.00) >DroYak_CAF1 160932 100 - 1 -------------GCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUUUUGCCUCGUUUUGCAAACAAAAAGCGUUGAGACCAACAA -------------(((..(((((((---(..((((.(((.......))).)))).))))).)))))).((((((((((.((........)))))))))))).((((....)))).. ( -31.20) >consensus _GCGGCAGCAGCAGCAAAGUCGGUU___GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUUUUGCCUCGUUUUGCAAACAAAAAGCGUUGAGACCAACAA .............(((..(((((((......((((.(((.......))).))))..)))).)))))).((((((((((.((........)))))))))))).((((....)))).. (-25.78 = -25.98 + 0.20)



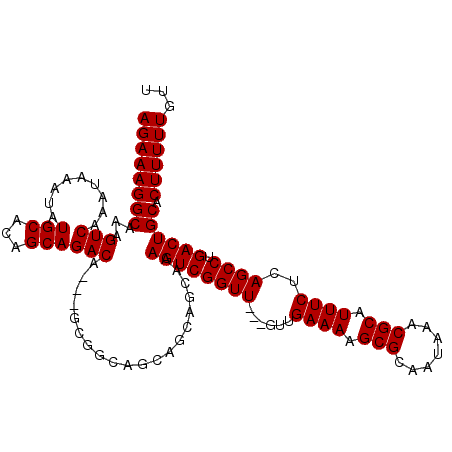
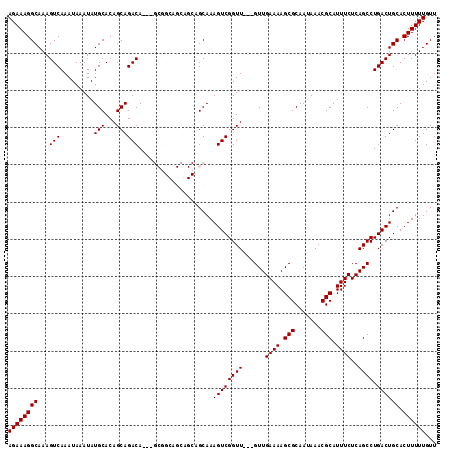
| Location | 5,048,166 – 5,048,275 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.29 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5048166 109 - 27905053 AGAAAGGCAAAGUCAAAUAAAUAUGCACAGCAGACA---GCGGCAGCAGCAGCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUU (((((((((..((((........(((...)))(((.---((.((....)).))...)))((((---(..((((.(((.......))).)))).)))))))))))).))))))... ( -29.20) >DroSec_CAF1 154439 109 - 1 AGAAAGGCAAAGUCAAAUAAAUAUGCACAGCAGACA---GCGGCAGCAGCAGCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUU (((((((((..((((........(((...)))(((.---((.((....)).))...)))((((---(..((((.(((.......))).)))).)))))))))))).))))))... ( -29.20) >DroSim_CAF1 160229 112 - 1 AGAAAGGCAAAGUCAAAUAAAUAUGCACAGCAGACAGCAGCGGCAGCAGCAGCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUU ((((((((...............(((...((.....)).((....)).)))....((((((((---(..((((.(((.......))).)))).))))).)))))).))))))... ( -29.70) >DroEre_CAF1 156825 100 - 1 AGAAAGGCAAAGUCAAAUAAAUAUGCACAGCAGACA---------------GCAAAGUCGGUUGUUGUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUU (((((((((..((((...........(((((.(((.---------------.....))).))))).(((((((((((.......))).))).))))).))))))).))))))... ( -24.40) >DroYak_CAF1 160971 97 - 1 AGAAAGGCAAAGUCAAAUAAAUAUGCACAGCAGACA---------------GCAAAGUCGGUU---GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUU (((((((((..((((........(((...)))(((.---------------.....)))((((---(..((((.(((.......))).)))).)))))))))))).))))))... ( -25.70) >consensus AGAAAGGCAAAGUCAAAUAAAUAUGCACAGCAGACA___GCGGCAGCAGCAGCAAAGUCGGUU___GUUGAAAAGCGCAAUAAACGCAUUUCUCAGCCUGACUGCACUUUUUGUU ((((((((...(((.........(((...))))))....................((((((((......((((.(((.......))).))))..)))).)))))).))))))... (-19.56 = -19.56 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:19 2006