
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,022,574 – 5,022,669 |
| Length | 95 |
| Max. P | 0.899454 |

| Location | 5,022,574 – 5,022,669 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -20.97 |
| Consensus MFE | -12.79 |
| Energy contribution | -14.35 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
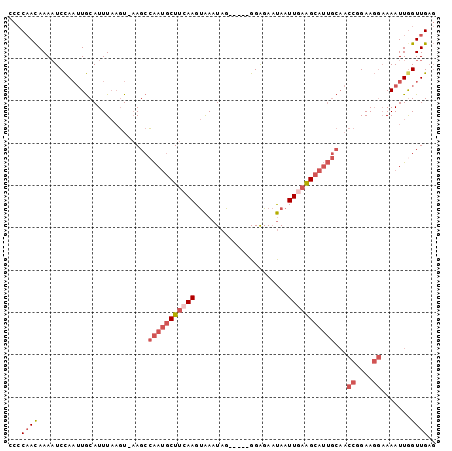
>3R_DroMel_CAF1 5022574 95 + 27905053 CCCCAACAAAAUCCAAUUGCAUUUAAGU-AAGCCAAUGCUUCAAGUAAAUAG-----GGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAAAAUUGGUUGAG ......(((...((((((..........-..((.((((((((((........-----.........))))))))))))..((....))..))))))))).. ( -23.13) >DroSec_CAF1 129144 95 + 1 CCCCAACAAAAUCCAAUUGCAUUUAAGU-AAGCCAAUGCUUCAAGUAAAUAG-----GGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAAAAUUAGUUGAG ...((((.........((((......))-))((.((((((((((........-----.........))))))))))))..((....)).......)))).. ( -22.63) >DroSim_CAF1 133592 95 + 1 CCCCAACAAAAUCCAAUUGCAUUUAAGU-AAGCCAAUGCUUCAAGUAAAUAG-----GGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAAAAUUGGUUGAG ......(((...((((((..........-..((.((((((((((........-----.........))))))))))))..((....))..))))))))).. ( -23.13) >DroEre_CAF1 130787 95 + 1 CCCCAACAAAAUCCAAUUGCAUUUAAGU-AAGCCAAUGCUUGAAGUAAAUAG-----GGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAAAAUUGGUUGAG ...((((....(((..((((.(((((((-(......))))))))))))....-----)))....................((....)).......)))).. ( -19.20) >DroYak_CAF1 133045 95 + 1 CCCCAACAAAAUCCAAUUGCAUUUAAGU-AAGCCAAUGCUUCAAGUAAAUAG-----GGGGAAUAAUUGAAGCAUUGCAACCGGAAGGAAAAUUGGUUGAG ......(((...((((((..........-..((.((((((((((........-----.........))))))))))))..((....))..))))))))).. ( -23.13) >DroAna_CAF1 128591 91 + 1 --ACUAUAAAUUCUAUAUACAUUUCUUUCUAGCAGACUCUCAAAGCGAAUAGAAAAAGGGGAUCAUUUGAGGCU--------UGAACUUAAAUUGGUUGCC --.......................((((((...(.((.....)))...))))))..((.(((((((((((...--------....)))))).))))).)) ( -14.60) >consensus CCCCAACAAAAUCCAAUUGCAUUUAAGU_AAGCCAAUGCUUCAAGUAAAUAG_____GGAGAAUAAUUGAAGCAUUGCAACCGGAAGGAAAAUUGGUUGAG ...((((..........................(((((((((((......................)))))))))))...((....)).......)))).. (-12.79 = -14.35 + 1.56)


| Location | 5,022,574 – 5,022,669 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -17.18 |
| Energy contribution | -18.74 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
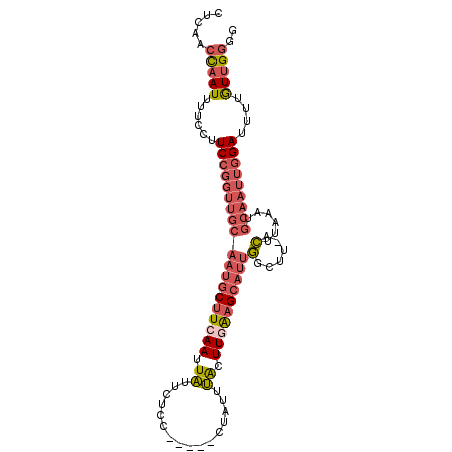
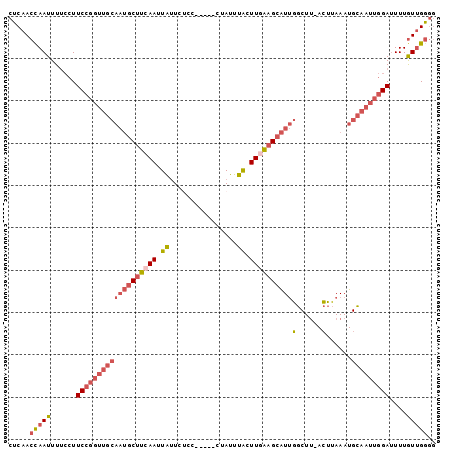
>3R_DroMel_CAF1 5022574 95 - 27905053 CUCAACCAAUUUUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCC-----CUAUUUACUUGAAGCAUUGGCUU-ACUUAAAUGCAAUUGGAUUUUGUUGGGG .....(((((......(((((((((((((((((((.((......-----.....)).))))))))))(....-.)......)))))))))....))))).. ( -27.00) >DroSec_CAF1 129144 95 - 1 CUCAACUAAUUUUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCC-----CUAUUUACUUGAAGCAUUGGCUU-ACUUAAAUGCAAUUGGAUUUUGUUGGGG ((((((..........(((((((((((((((((((.((......-----.....)).))))))))))(....-.)......)))))))))....)))))). ( -25.94) >DroSim_CAF1 133592 95 - 1 CUCAACCAAUUUUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCC-----CUAUUUACUUGAAGCAUUGGCUU-ACUUAAAUGCAAUUGGAUUUUGUUGGGG .....(((((......(((((((((((((((((((.((......-----.....)).))))))))))(....-.)......)))))))))....))))).. ( -27.00) >DroEre_CAF1 130787 95 - 1 CUCAACCAAUUUUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCC-----CUAUUUACUUCAAGCAUUGGCUU-ACUUAAAUGCAAUUGGAUUUUGUUGGGG .....(((((......((((((((((((((((.((.((......-----.....)).)).)))))))(....-.)......)))))))))....))))).. ( -20.80) >DroYak_CAF1 133045 95 - 1 CUCAACCAAUUUUCCUUCCGGUUGCAAUGCUUCAAUUAUUCCCC-----CUAUUUACUUGAAGCAUUGGCUU-ACUUAAAUGCAAUUGGAUUUUGUUGGGG .....(((((......(((((((((((((((((((.((......-----.....)).))))))))))(....-.)......)))))))))....))))).. ( -27.00) >DroAna_CAF1 128591 91 - 1 GGCAACCAAUUUAAGUUCA--------AGCCUCAAAUGAUCCCCUUUUUCUAUUCGCUUUGAGAGUCUGCUAGAAAGAAAUGUAUAUAGAAUUUAUAGU-- (....).....(((((((.--------...(((((((((..............))).))))))..(((.......)))..........)))))))....-- ( -11.14) >consensus CUCAACCAAUUUUCCUUCCGGUUGCAAUGCUUCAAUUAUUCUCC_____CUAUUUACUUGAAGCAUUGGCUU_ACUUAAAUGCAAUUGGAUUUUGUUGGGG .....(((((......(((((((((((((((((((.((................)).))))))))))(......)......)))))))))....))))).. (-17.18 = -18.74 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:10 2006