
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,008,073 – 5,008,171 |
| Length | 98 |
| Max. P | 0.875749 |

| Location | 5,008,073 – 5,008,171 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -11.83 |
| Energy contribution | -12.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
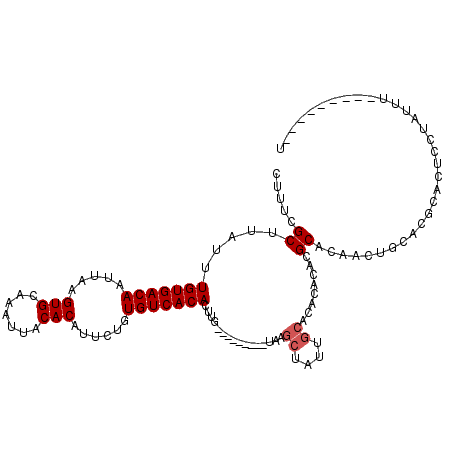
>3R_DroMel_CAF1 5008073 98 + 27905053 CUUUCGCUUAUUUGUGACAAUUAAGUGCAAAUUACACAUUCUGUGUCACACUUG---------UAAGCUAUUGCAAACACCCGCACAACUGAACGCACUCUUAUUU---------U ..(((((((((.(((((((.....(((.......)))......)))))))...)---------)))))...(((........))).....))).............---------. ( -19.30) >DroSec_CAF1 114695 97 + 1 CUUUCGCUUAUUUGUGACAAUUAAGUGCAAAUUACACAUUCUGUGUCACACUUG---------UAAACUAUUGCACACACAAGCACA-CUGCACGCACUCCUAUUU---------U .....((((.(.((((.((((....(((((...((((.....)))).....)))---------))....)))))))).).))))...-..................---------. ( -17.70) >DroSim_CAF1 118614 98 + 1 CUUUCGCUUAUUUGUGACAAUUAAGUGCAAAUUACACAUUCUGUGUCACACUUG---------UAAGCUAUUGCACACACAAGCACAACUGCACGCACUCCUAUUU---------U .....((((((.(((((((.....(((.......)))......)))))))...)---------)))))..............(((....)))..............---------. ( -19.10) >DroEre_CAF1 116222 107 + 1 CUUUCGCUUAUUUGUGACAAUUAAGUGCAAAUUACACAUUCUGUGUCACACUUG---------UAAGCUAUUGCACACAUUCGCACAACUGCACGCACUCUUACUGUGUGCUGUGU .....((((((.(((((((.....(((.......)))......)))))))...)---------)))))..............(((((...(((((((.......)))))))))))) ( -30.10) >DroYak_CAF1 117413 107 + 1 CUUUCGCUUAUUUGUGACAAUUAAGUGCAAAUUACACAUUCUGUGUCACACUUGUGCAAGUAGUAAGCUAUUGCACACACCCGCACUGCUGCACGCACUCGUACUG---------A ...((((......))))......(((((.....((((.....)))).......(((((.(((((..((...((....))...))))))))))))))))).......---------. ( -25.30) >DroAna_CAF1 110746 79 + 1 CUUUCGCUUAUUUGUGACAAUUAAGUGCAAAUUACACAUUCUGUGUCACACUUG---------UCGGGUCAAACACACCCACGCA-------------------UA---------U .....((.....(((((((.....(((.......)))......)))))))...(---------(.((((.......)))))))).-------------------..---------. ( -17.60) >consensus CUUUCGCUUAUUUGUGACAAUUAAGUGCAAAUUACACAUUCUGUGUCACACUUG_________UAAGCUAUUGCACACACACGCACAACUGCACGCACUCCUAUUU_________U .....((.....(((((((.....(((.......)))......)))))))................((....))........))................................ (-11.83 = -12.17 + 0.33)

| Location | 5,008,073 – 5,008,171 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.98 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5008073 98 - 27905053 A---------AAAUAAGAGUGCGUUCAGUUGUGCGGGUGUUUGCAAUAGCUUA---------CAAGUGUGACACAGAAUGUGUAAUUUGCACUUAAUUGUCACAAAUAAGCGAAAG .---------(((((...(..((......))..)...)))))......(((((---------....(((((((...((.((((.....))))))...)))))))..)))))..... ( -24.20) >DroSec_CAF1 114695 97 - 1 A---------AAAUAGGAGUGCGUGCAG-UGUGCUUGUGUGUGCAAUAGUUUA---------CAAGUGUGACACAGAAUGUGUAAUUUGCACUUAAUUGUCACAAAUAAGCGAAAG .---------........(..((..(((-.....)))..))..)....(((((---------....(((((((...((.((((.....))))))...)))))))..)))))..... ( -24.70) >DroSim_CAF1 118614 98 - 1 A---------AAAUAGGAGUGCGUGCAGUUGUGCUUGUGUGUGCAAUAGCUUA---------CAAGUGUGACACAGAAUGUGUAAUUUGCACUUAAUUGUCACAAAUAAGCGAAAG .---------........(..((..(((......)))..))..)....(((((---------....(((((((...((.((((.....))))))...)))))))..)))))..... ( -27.30) >DroEre_CAF1 116222 107 - 1 ACACAGCACACAGUAAGAGUGCGUGCAGUUGUGCGAAUGUGUGCAAUAGCUUA---------CAAGUGUGACACAGAAUGUGUAAUUUGCACUUAAUUGUCACAAAUAAGCGAAAG .....(((((((......(..((......))..)...)))))))....(((((---------....(((((((...((.((((.....))))))...)))))))..)))))..... ( -33.60) >DroYak_CAF1 117413 107 - 1 U---------CAGUACGAGUGCGUGCAGCAGUGCGGGUGUGUGCAAUAGCUUACUACUUGCACAAGUGUGACACAGAAUGUGUAAUUUGCACUUAAUUGUCACAAAUAAGCGAAAG (---------(.(((((....))))).((.(((((((((((.((....)).))).))))))))...(((((((...((.((((.....))))))...))))))).....))))... ( -36.10) >DroAna_CAF1 110746 79 - 1 A---------UA-------------------UGCGUGGGUGUGUUUGACCCGA---------CAAGUGUGACACAGAAUGUGUAAUUUGCACUUAAUUGUCACAAAUAAGCGAAAG .---------..-------------------(((((((((.......)))).)---------)...(((((((...((.((((.....))))))...))))))).....))).... ( -21.00) >consensus A_________AAAUAAGAGUGCGUGCAGUUGUGCGUGUGUGUGCAAUAGCUUA_________CAAGUGUGACACAGAAUGUGUAAUUUGCACUUAAUUGUCACAAAUAAGCGAAAG ...................((((..((..........))..))))...(((((.............(((((((...((.((((.....))))))...)))))))..)))))..... (-15.61 = -16.98 + 1.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:08 2006