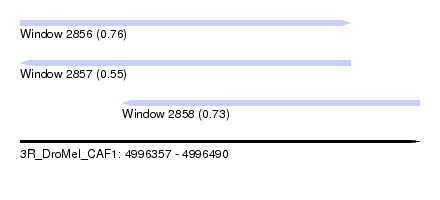
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,996,357 – 4,996,490 |
| Length | 133 |
| Max. P | 0.760464 |

| Location | 4,996,357 – 4,996,467 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4996357 110 + 27905053 AUUAAAGCCAAGACAAACAAACUC-----GCAAUGGGGC----GCAGACACGUGCAAAAUGAGU-UAACAAUUUGCAUGCAAAUCUGCGCCGACAAGUCGCCUAAUGUUAACCCAUUAAG ........................-----..((((((((----(((((..((((((((.((...-...)).))))))))....)))))))(((....)))...........))))))... ( -31.00) >DroPse_CAF1 146904 103 + 1 ---AAAGC-----------ACUUCGAGGGGCAG-GGGGAGGGGCUAGACACGUGCAAAAUGAGU-UAACAAUUUGCAUGUUAAUUUGCUCCGACAAGUCGCUUAAUGUUAACCCAU-GAA ---.....-----------...(((.((((((.-.(((.(((((.....(((((((((.((...-...)).)))))))))......)))))((....)).)))..)))...))).)-)). ( -29.40) >DroSim_CAF1 106883 110 + 1 AUUAAAGCCAAGACAAACAAACUC-----GCAAUGGGGC----GCAGACACGUGCAAAAUGAGU-UAACAAUUUGCAUGCAAAUCUGCGCCGACAAGUCGCCUAAUGUUAACCCAUUAAG ........................-----..((((((((----(((((..((((((((.((...-...)).))))))))....)))))))(((....)))...........))))))... ( -31.00) >DroEre_CAF1 103998 110 + 1 AUUAAAGCCAAGACAAACAAACUC-----GCAAUGGGGC----GCAGACACGUGCAAAAUGAGU-UAACAAUUUGCAUGCAAAUCUGCGUCGACAAGUCGCCUAAUGUUAACCCAUUAAG ........................-----..((((((((----(((((..((((((((.((...-...)).))))))))....)))))))(((....)))...........))))))... ( -29.10) >DroYak_CAF1 105353 110 + 1 AUUAAAGCCAAGACAAACAAACUC-----GCAAUGGGGC----GCAGACACGUGCAAAAUGAGU-UAACAAUUUGCAUGCAAAUCUGCGCCGACAAGUCGCCUAAUGUUAACCCAUUAAG ........................-----..((((((((----(((((..((((((((.((...-...)).))))))))....)))))))(((....)))...........))))))... ( -31.00) >DroAna_CAF1 97868 113 + 1 ACUACUGACAAGACAAACA--UUC-----ACACUGGGUUGCUGGCAGACACGUGCAAAAUGAGUUUAACAAUUUGUAUGUAAAUCCGCGCCGACAAGUCGCCUAAUGUUAACCCAUUAAG ...................--...-----....((((((((.(((.((((((((((((.((.......)).)))))))))...((......))...))))))....).)))))))..... ( -23.70) >consensus AUUAAAGCCAAGACAAACAAACUC_____GCAAUGGGGC____GCAGACACGUGCAAAAUGAGU_UAACAAUUUGCAUGCAAAUCUGCGCCGACAAGUCGCCUAAUGUUAACCCAUUAAG ...............................((((((......(((((..((((((((.((.......)).))))))))....)))))..(((....)))...........))))))... (-21.38 = -21.68 + 0.31)



| Location | 4,996,357 – 4,996,467 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.87 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4996357 110 - 27905053 CUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCAGAUUUGCAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUGC----GCCCCAUUGC-----GAGUUUGUUUGUCUUGGCUUUAAU ......(((((((...((((((((((((.(((((((((..((.((((((.((...-...)).)))))).))))))))----))).....))-----))).)))))))..))))))).... ( -37.10) >DroPse_CAF1 146904 103 - 1 UUC-AUGGGUUAACAUUAAGCGACUUGUCGGAGCAAAUUAACAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUAGCCCCUCCCC-CUGCCCCUCGAAGU-----------GCUUU--- ...-(((((((((((.....(((....)))..(((........)))....)))))-))))))...((((...((.((...(.....-..)...)).)).))-----------))...--- ( -21.50) >DroSim_CAF1 106883 110 - 1 CUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCAGAUUUGCAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUGC----GCCCCAUUGC-----GAGUUUGUUUGUCUUGGCUUUAAU ......(((((((...((((((((((((.(((((((((..((.((((((.((...-...)).)))))).))))))))----))).....))-----))).)))))))..))))))).... ( -37.10) >DroEre_CAF1 103998 110 - 1 CUUAAUGGGUUAACAUUAGGCGACUUGUCGACGCAGAUUUGCAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUGC----GCCCCAUUGC-----GAGUUUGUUUGUCUUGGCUUUAAU (((((((......)))))))(((....))).(((((((..((.((((((.((...-...)).)))))).))))))))----)..(((..((-----(((....)))))..)))....... ( -31.50) >DroYak_CAF1 105353 110 - 1 CUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCAGAUUUGCAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUGC----GCCCCAUUGC-----GAGUUUGUUUGUCUUGGCUUUAAU ......(((((((...((((((((((((.(((((((((..((.((((((.((...-...)).)))))).))))))))----))).....))-----))).)))))))..))))))).... ( -37.10) >DroAna_CAF1 97868 113 - 1 CUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCGGAUUUACAUACAAAUUGUUAAACUCAUUUUGCACGUGUCUGCCAGCAACCCAGUGU-----GAA--UGUUUGUCUUGUCAGUAGU .(((.((((((.......(.(((....))).)((((((..((...((((.((.......)).))))...))))))))....))))))...)-----)).--................... ( -21.80) >consensus CUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCAGAUUUGCAUGCAAAUUGUUA_ACUCAUUUUGCACGUGUCUGC____GCCCCAUUGC_____GAGUUUGUUUGUCUUGGCUUUAAU ..(((((((.........(.(((....))).)((((((..((.((((((.((.......)).)))))).))))))))......))))))).............................. (-21.86 = -22.87 + 1.00)



| Location | 4,996,391 – 4,996,490 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -24.98 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4996391 99 - 27905053 -----------------CAGCGACGACAGCUUUUCACGGCCUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCAGAUUUGCAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUGC--- -----------------...((((((..((((.....(((((....)))))......))))...))))))..((((((..((.((((((.((...-...)).)))))).))))))))--- ( -30.90) >DroPse_CAF1 146929 101 - 1 -----------------CAACGACGACAGCUUUUCACGGCUUC-AUGGGUUAACAUUAAGCGACUUGUCGGAGCAAAUUAACAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUAGCCC -----------------...........(((...((((((...-(((((((((((.....(((....)))..(((........)))....)))))-))))))...)).))))...))).. ( -29.20) >DroEre_CAF1 104032 99 - 1 -----------------CAGCGACGACAGCUUUUCACGGCCUUAAUGGGUUAACAUUAGGCGACUUGUCGACGCAGAUUUGCAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUGC--- -----------------..(((((((..((((.....(((((....)))))......))))...)))))).)((((((..((.((((((.((...-...)).)))))).))))))))--- ( -30.90) >DroYak_CAF1 105387 99 - 1 -----------------CAGCGACGACAGCUUUUCACGGCCUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCAGAUUUGCAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUGC--- -----------------...((((((..((((.....(((((....)))))......))))...))))))..((((((..((.((((((.((...-...)).)))))).))))))))--- ( -30.90) >DroAna_CAF1 97901 120 - 1 GCAGAAACUGGCAGCAACAGCGACAGAAGCUUUUCAGGGCCUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCGGAUUUACAUACAAAUUGUUAAACUCAUUUUGCACGUGUCUGCCAG .......(((((((((...((((.....(((...((((((((.((((......))))))))..))))..)))..(..((((((.......)).))))..)...))))...)).))))))) ( -33.60) >DroPer_CAF1 152822 102 - 1 -----------------CAACGACGACAGCUUUUCACGGCUUUAAUGGGUUAACAUUAAGCGACUUGUCGGAGCAAAUUUACAUGCAAAUUGUUA-ACUCAUUUUGCACGUGUCUAGCCC -----------------...........(((...((((((...((((((((((((.....(((....)))..(((........)))....)))))-)))))))..)).))))...))).. ( -30.50) >consensus _________________CAGCGACGACAGCUUUUCACGGCCUUAAUGGGUUAACAUUAGGCGACUUGUCGGCGCAGAUUUACAUGCAAAUUGUUA_ACUCAUUUUGCACGUGUCUGC___ ....................((((((..((((.....(((((....)))))......))))...))))))..((((((..((.((((((.((.......)).)))))).))))))))... (-24.98 = -25.03 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:06 2006