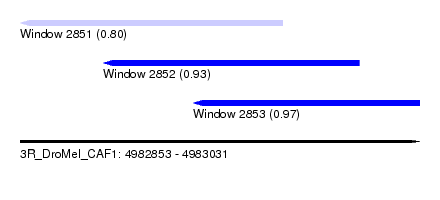
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,982,853 – 4,983,031 |
| Length | 178 |
| Max. P | 0.966272 |

| Location | 4,982,853 – 4,982,970 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -34.53 |
| Energy contribution | -32.57 |
| Covariance contribution | -1.96 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
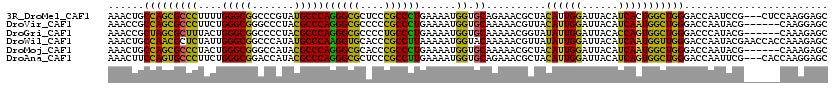
>3R_DroMel_CAF1 4982853 117 - 27905053 AAACUGCCAGCGCCCUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAGAAACGCUACAUUGGAUUACAUCACUGGCUGGGACCAAUCCG---CUCCAAGGAGC ...(.((((((((((....)))))....(((..(((((((((....)))))))..((((..((......))..))))))..)))....))))).)(((....)))(---(((....)))) ( -46.50) >DroVir_CAF1 116151 114 - 1 AAACCGCCAGCGCCCUUCUGGGCGGGCCCUACGCCCAGGGCGCCCCCGCCCUGAAAAUGGUGCAAAAACGUUACAUUGGAUUACAUCAAUGGCUGGGACCAAUACG------CAAGGAGC ......((((((((((((.(((((((((((......)))))....)))))).)))...)).))..........((((((......)))))))))))........(.------...).... ( -41.30) >DroGri_CAF1 110633 114 - 1 AAACCGCUAGCGCUUUACUGGGCGGCCCCUACGCCCAGGGCGCCCCUGCCCUGAAAAUGGUGCAAAAACGGUAUAUUGGAUUACACCAGUGGCUGGGACCCAUACG------CAAAGAGC .....((..(((((...(((((((.......))))))))))))(((.((((((......((......)).(((........)))..))).))).)))........)------)....... ( -39.40) >DroWil_CAF1 98359 120 - 1 AAACUGCCAACGCUCUAUUGGGCGGCCCAUAUGCCCAAGGUGCACCCGCCUUAAAAAUGGUACAAAAACGUUAUAUUGGAUUACAUCAAUGGUUGGGACCAAUACGAACCACCAAAGAGC ...........(((((.(((((((.......)))))))((((.....(((........)))........(((.((((((.(..((........))..)))))))..)))))))..))))) ( -31.20) >DroMoj_CAF1 106745 114 - 1 AAACUGCCAGCGCCCUACUGGGCGGGCCAUACGCCCAGGGCGCACCCGCCCUGAAAAUGGUGCAAAAACGCUACAUUGGAUUACAUCAAUGGCUGGGACCAAUACG------CAAAGAGC ....(((..(((((((...(((((.......)))))))))))).(((((((.......)).))......((..((((((......)))))))).)))........)------))...... ( -41.20) >DroAna_CAF1 85029 117 - 1 AAACUUCCAGUGCCCUUCUGGGCGGACCAUACGCCCAGGGCGCUCCCGCCUUGAAAAUGGUGCAGAAACGCUACAUUGGAUUACAUCAGUGGCUGGGACCAAUUCG---CACCAAGGAGC ....((((((((((((...(((((.......))))))))))))).............((((((.(((..(((((..((.....))...)))))((....)).))))---))))).)))). ( -45.10) >consensus AAACUGCCAGCGCCCUACUGGGCGGCCCAUACGCCCAGGGCGCACCCGCCCUGAAAAUGGUGCAAAAACGCUACAUUGGAUUACAUCAAUGGCUGGGACCAAUACG______CAAAGAGC ......(((((((((....(((((.......)))))((((((....))))))......)).))..........((((((......)))))))))))........................ (-34.53 = -32.57 + -1.96)

| Location | 4,982,890 – 4,983,004 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -36.23 |
| Energy contribution | -35.23 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4982890 114 - 27905053 GCAGAUCCGGGCGCU---AGC---AAAAUGGUGGUUUUAGAAACUGCCAGCGCCCUUUUGGGCGGCCCGUAUGCCCAGGGCGCUCCCGCCCUGAAAAUGGUGCAGAAACGCUACAUUGGA (((....(((((...---...---....(((..(((.....)))..))).(((((....))))))))))..))).(((((((....)))))))..((((..((......))..))))... ( -48.40) >DroVir_CAF1 116185 117 - 1 GGCACCACAAGCGAUUACAAC---AAAAUGGUGGUUUUAGAAACCGCCAGCGCCCUUCUGGGCGGGCCCUACGCCCAGGGCGCCCCCGCCCUGAAAAUGGUGCAAAAACGUUACAUUGGA .((((((...(((........---....((((((((.....)))))))).(((((....))))).......))).(((((((....)))))))....))))))................. ( -48.70) >DroGri_CAF1 110667 117 - 1 GGCACAACAAGCGAUUACAAC---AAAAUGGUGGUUCUGGAAACCGCUAGCGCUUUACUGGGCGGCCCCUACGCCCAGGGCGCCCCUGCCCUGAAAAUGGUGCAAAAACGGUAUAUUGGA .((((...(((((........---....((((((((.....)))))))).)))))....(((((.......)))))((((((....)))))).......))))................. ( -37.72) >DroWil_CAF1 98399 113 - 1 ACAGAAACA-ACAGU---AGC---AAAAUGGUGGUUUUAGAAACUGCCAACGCUCUAUUGGGCGGCCCAUAUGCCCAAGGUGCACCCGCCUUAAAAAUGGUACAAAAACGUUAUAUUGGA .......((-(..((---(((---....(((..(((.....)))..)))..(..((((((((((.......)))))((((((....))))))...)))))..)......))))).))).. ( -29.30) >DroMoj_CAF1 106779 117 - 1 GGGACCGCAAGCGAUUUCAAC---AAAAUGGUGGUUUUAGAAACUGCCAGCGCCCUACUGGGCGGGCCAUACGCCCAGGGCGCACCCGCCCUGAAAAUGGUGCAAAAACGCUACAUUGGA ....(((..((((........---....(((..(((.....)))..)))(((((.....(((((.......)))))((((((....))))))......))))).....))))....))). ( -44.40) >DroPer_CAF1 134665 117 - 1 ACAAGCCCAAGCGAU---AGCCACAAAAUGGUGGUUCUAGAGACUGCCAGCGCCCUGCUGGGCGGCCCAUAUGCCCAGGGCGCGCCCGCCUUGAAAAUGGUACAAAAACGCUACAUUGGA ......(((((((..---.((.......(((..(((.....)))..)))(((((((...(((((.......))))))))))))))..(((........))).......))))....))). ( -41.70) >consensus GCAACCACAAGCGAU___AAC___AAAAUGGUGGUUUUAGAAACUGCCAGCGCCCUACUGGGCGGCCCAUACGCCCAGGGCGCACCCGCCCUGAAAAUGGUGCAAAAACGCUACAUUGGA .........((((...............((((((((.....))))))))(((((.....(((((.......)))))((((((....))))))......))))).....))))........ (-36.23 = -35.23 + -0.99)

| Location | 4,982,930 – 4,983,031 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.02 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -21.83 |
| Energy contribution | -20.94 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
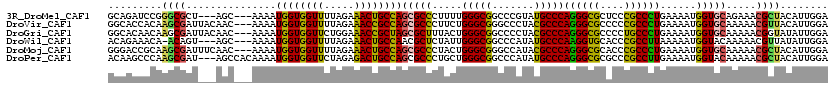
>3R_DroMel_CAF1 4982930 101 - 27905053 AAGCAUCAACUAUCAACAGCAACUAUCGCAGAUCCGGGCGCU---AGC---AAAAUGGUGGUUUUAGAAACUGCCAGCGCCCUUUUGGGCGGCCCGUAUGCCCAGGG ..((((............((.......)).....(((((...---...---....(((..(((.....)))..))).(((((....)))))))))).))))...... ( -32.60) >DroPse_CAF1 128207 97 - 1 ----AACAACA---CACAACAGCUAUCACAGGCCGAAGCGAU---AGCCACAAAAUGGUGGUUCUAGAGACUGCCAGCGCCCUGCUGGGCGGCCCAUAUGCCCAGGG ----.......---..............((((.((..((...---.)).......(((..(((.....)))..))).)).))))(((((((.......))))))).. ( -28.70) >DroYak_CAF1 91634 95 - 1 ------CAACUAGCAACAGCAACUAUCGCUGAUCCGGGCGCU---AGC---AAAAUGGUGGUUUUAGAAACUACCACCGCCCUUUUGGGCGGCCCCUAUGCCCAAGG ------..........((((.......))))....(((((.(---((.---....((((((((.....))))))))((((((....))))))...)))))))).... ( -37.10) >DroMoj_CAF1 106819 99 - 1 ----AACAACA-AAAACUAAAAUAAUCGGGACCGCAAGCGAUUUCAAC---AAAAUGGUGGUUUUAGAAACUGCCAGCGCCCUACUGGGCGGGCCAUACGCCCAGGG ----.......-...............(((..(((....(....)...---....(((..(((.....)))..)))))))))..(((((((.......))))))).. ( -30.00) >DroAna_CAF1 85106 94 - 1 ---CUGCAGCU----AUAACUUCGUUUGCAGACCCGGGCGAU---AGC---AAAAUGGUGGUUUUAGAAACUUCCAGUGCCCUUCUGGGCGGACCAUACGCCCAGGG ---(((((((.----........).)))))).((((((((..---...---....(((.((((.....)))).))).(((((....))))).......))))).))) ( -33.30) >DroPer_CAF1 134705 97 - 1 ----AACAACA---CACAACAGCUAUCACAAGCCCAAGCGAU---AGCCACAAAAUGGUGGUUCUAGAGACUGCCAGCGCCCUGCUGGGCGGCCCAUAUGCCCAGGG ----.......---.......((((((.(........).)))---))).......(((..(((.....)))..))).(((((....))))).(((.........))) ( -28.30) >consensus ____AACAACA___AACAACAACUAUCGCAGACCCAAGCGAU___AGC___AAAAUGGUGGUUUUAGAAACUGCCAGCGCCCUUCUGGGCGGCCCAUAUGCCCAGGG .....................................(((...............((((((((.....)))))))).)))....(((((((.......))))))).. (-21.83 = -20.94 + -0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:01 2006