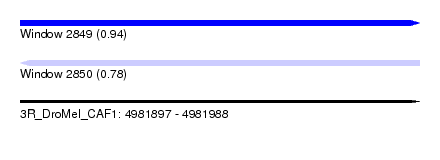
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,981,897 – 4,981,988 |
| Length | 91 |
| Max. P | 0.941130 |

| Location | 4,981,897 – 4,981,988 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.87 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4981897 91 + 27905053 --CACUCUAUUUCUC--UGCUCCUUCCAC--AUUCGGUUCUGGCCCACAGCAUAAUUCAUAUGCUAUUUAAGCCCGGGCCAUAAAUUUACCGACCAA --.............--............--..(((((..((((((..((((((.....))))))..........)))))).......))))).... ( -18.60) >DroPse_CAF1 127707 97 + 1 AUCGCUCUCACCCAGACCGCCGCAUCCACUCGCUCGGCUCUGGCCCAAAGCAUAAUUCAUAUGCUGCUUAAGCCUGGGCCAUAAAUUUACCGACCAA .(((.........((((((..((........)).))).)))((((((.((((((.....))))))((....)).))))))..........))).... ( -25.80) >DroEre_CAF1 89432 91 + 1 --CACUCUAUUCUUG--CACUUCUUCCAC--AUUCGGUUCUGGCCCAAAGCAUAAUUCAUAUGCUAUUUAAGCCCGGGCCAUAAAUUUACCGACUAA --.............--............--..(((((..((((((..((((((.....))))))..........)))))).......))))).... ( -18.60) >DroYak_CAF1 90603 91 + 1 --CACUCUAUUCCCG--CUCGCCUUCCAC--UUUCGGUUCUGGCCCAAAGCAUAAUUCAUAUGCUAUUUAAGCCCGGGCCAUAAAUUUACCGACCAA --.............--............--..(((((..((((((..((((((.....))))))..........)))))).......))))).... ( -18.60) >DroAna_CAF1 84068 93 + 1 CUCGCUCUAAUUCGG--UUUUCCUCCCAC--UCUCGGUUCUGCCCCAGAGCAUAAUUCAUAUGCUAUUUAAGCCUGGGCCAUAAAUUCACCGGCCAA ...........((((--(.......((..--....))...((.(((((((((((.....))))))........))))).)).......))))).... ( -17.40) >DroPer_CAF1 134207 97 + 1 AUCGCUCUCACCCAGACCGCCGCAUCCACUCGCUCGGCUCUGGCCCAAAGCAUAAUUCAUAUGCUGCUUAAGCCUGGGCCAUAAAUUUACCGACCAA .(((.........((((((..((........)).))).)))((((((.((((((.....))))))((....)).))))))..........))).... ( -25.80) >consensus __CACUCUAAUCCAG__CGCUCCUUCCAC__ACUCGGUUCUGGCCCAAAGCAUAAUUCAUAUGCUAUUUAAGCCCGGGCCAUAAAUUUACCGACCAA ...................................(((((.(((....((((((.....))))))......))).)))))................. (-16.92 = -16.87 + -0.06)


| Location | 4,981,897 – 4,981,988 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -22.03 |
| Energy contribution | -21.95 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4981897 91 - 27905053 UUGGUCGGUAAAUUUAUGGCCCGGGCUUAAAUAGCAUAUGAAUUAUGCUGUGGGCCAGAACCGAAU--GUGGAAGGAGCA--GAGAAAUAGAGUG-- ....(((((.......((((((........((((((((.....))))))))))))))..))))).(--((.......)))--.............-- ( -23.80) >DroPse_CAF1 127707 97 - 1 UUGGUCGGUAAAUUUAUGGCCCAGGCUUAAGCAGCAUAUGAAUUAUGCUUUGGGCCAGAGCCGAGCGAGUGGAUGCGGCGGUCUGGGUGAGAGCGAU (((.(((((.......((((((((((....))((((((.....))))))))))))))..))))).)))..........((.(((.....))).)).. ( -34.30) >DroEre_CAF1 89432 91 - 1 UUAGUCGGUAAAUUUAUGGCCCGGGCUUAAAUAGCAUAUGAAUUAUGCUUUGGGCCAGAACCGAAU--GUGGAAGAAGUG--CAAGAAUAGAGUG-- ....(((((.......((((((((........((((((.....))))))))))))))..)))))..--............--.............-- ( -22.70) >DroYak_CAF1 90603 91 - 1 UUGGUCGGUAAAUUUAUGGCCCGGGCUUAAAUAGCAUAUGAAUUAUGCUUUGGGCCAGAACCGAAA--GUGGAAGGCGAG--CGGGAAUAGAGUG-- ............(((((..(((((((((((..((((((.....)))))))))))))....(((...--.)))........--)))).)))))...-- ( -25.20) >DroAna_CAF1 84068 93 - 1 UUGGCCGGUGAAUUUAUGGCCCAGGCUUAAAUAGCAUAUGAAUUAUGCUCUGGGGCAGAACCGAGA--GUGGGAGGAAAA--CCGAAUUAGAGCGAG ..(((((.........)))))...((((.(((.(((((.....)))))(((.((......)).)))--.(((........--))).))).))))... ( -24.00) >DroPer_CAF1 134207 97 - 1 UUGGUCGGUAAAUUUAUGGCCCAGGCUUAAGCAGCAUAUGAAUUAUGCUUUGGGCCAGAGCCGAGCGAGUGGAUGCGGCGGUCUGGGUGAGAGCGAU (((.(((((.......((((((((((....))((((((.....))))))))))))))..))))).)))..........((.(((.....))).)).. ( -34.30) >consensus UUGGUCGGUAAAUUUAUGGCCCAGGCUUAAAUAGCAUAUGAAUUAUGCUUUGGGCCAGAACCGAAA__GUGGAAGGAGCG__CAGGAAUAGAGCG__ ....(((((.......((((((((........((((((.....))))))))))))))..)))))................................. (-22.03 = -21.95 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:58 2006