
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,972,489 – 4,972,693 |
| Length | 204 |
| Max. P | 0.974850 |

| Location | 4,972,489 – 4,972,586 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.90 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.77 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4972489 97 - 27905053 CCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCCAAACGUGCCGCCAUGUGAGGCGCAUAUGAAAACUGUCAAAGUGUAUGGAAGAUACCA ((((((...(((((..(((.......)))....(((((((..(((((....)))))..))).)))).......)))))...)))).))......... ( -24.70) >DroSec_CAF1 79328 97 - 1 CCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCCAAACGUGCCGCCAUGUGAGGCGCGUUUGAAAAAUGUCAAAGUGUAUGGAAGAUACCA ((((((...(((((..(((.......))).(((...)))((((((((((.........)))))))))).....)))))...)))).))......... ( -30.70) >DroSim_CAF1 81140 97 - 1 CCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCCAAACGUGCCGCCAUGUGAGGCGCGUUUGAAAACUGUCAAAGUGUAUGGAAGAUACCA ((((((...(((((..(((.......))).(((...)))((((((((((.........)))))))))).....)))))...)))).))......... ( -30.50) >DroEre_CAF1 80015 97 - 1 CCUACAAACUGACAAAGAGUCAUCUUUUCAGCCUAUAGCCAAACGUGCCGCCAUGUGAGGAGCGUUUGAAAACUGUCAAAGUAUAUGGAAGAUACCA ........((((.(((((....))))))))).(((((..(((((((.((.........)).)))))))...(((.....))).)))))......... ( -20.30) >DroYak_CAF1 81032 97 - 1 CCUACAAACUGACAAAGAGUCAACUUUUCAGCCUAUAGCCAAACGUGCCGCCAUGUGAGGCGCGUUUGAAAACUGUCAAAGUAUAUGGAAGAUACCA ........((((.((((......)))))))).(((((..((((((((((.........))))))))))...(((.....))).)))))......... ( -24.00) >DroAna_CAF1 75373 86 - 1 C-------CUGACAAAAAGUCAACUU--CAGCCUAUGGCCAAACGUGCUGCCAU--GAAGCGCGUUUGAAAAGUGUCAAAGUAUCUAGCAGAUACAC .-------.(((((............--..(((...)))((((((((((.....--..)))))))))).....)))))..(((((.....))))).. ( -26.90) >consensus CCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCCAAACGUGCCGCCAUGUGAGGCGCGUUUGAAAACUGUCAAAGUAUAUGGAAGAUACCA .........(((((................((.....))((((((((((.........)))))))))).....)))))..((((.......)))).. (-20.79 = -20.77 + -0.03)



| Location | 4,972,515 – 4,972,618 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4972515 103 - 27905053 AAAGUAUCUGACAGAUACAUAUG--------CUCGUGCUGCCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCCAAACGUGCCGCCAU------GUGAGGCGCAUAUGAAA--- ...(((((.....)))))(((((--------(..(.((((.........((((.....))))......)))))....(((..(((((....)))------))..)))))))))....--- ( -27.06) >DroPse_CAF1 118361 107 - 1 ---GUAUCUGACAGAUACAUUUG--------CUAGCGAUGCCUACAAACUGACAAAAAGUCAACUC--CAGCCUAGGCCCGGACGUGCCACAGUGCCGCCGCGUGGCGCAUUUGAAAAGA ---(((((.....))))).((((--------.(((......))))))).((((.....))))..((--(.((....))..))).(((((((.(((....))))))))))........... ( -28.40) >DroEre_CAF1 80041 103 - 1 AAAGUAUCUGACAGAUACAUAUG--------CUCGUGCUGCCUACAAACUGACAAAGAGUCAUCUUUUCAGCCUAUAGCCAAACGUGCCGCCAU------GUGAGGAGCGUUUGAAA--- ...(((((.....)))))..(((--------((((.((((........((((.(((((....)))))))))....)))))..(((((....)))------))...))))))......--- ( -23.40) >DroYak_CAF1 81058 103 - 1 AAAGUAUCUGACAGAUACAUAUG--------CUCGUGCUGCCUACAAACUGACAAAGAGUCAACUUUUCAGCCUAUAGCCAAACGUGCCGCCAU------GUGAGGCGCGUUUGAAA--- ...(((((.....)))))....(--------((...((((.........((((.....))))......))))....)))((((((((((.....------....))))))))))...--- ( -29.26) >DroAna_CAF1 75399 100 - 1 CAAGUAUCUGACAGAUACAUAUGCCGGCAUGCUCCUGCUGC-------CUGACAAAAAGUCAACUU--CAGCCUAUGGCCAAACGUGCUGCCAU--------GAAGCGCGUUUGAAA--- ...(((((.....)))))....(((((((.((....)))))-------)((((.....))))....--........)))((((((((((.....--------..))))))))))...--- ( -35.80) >DroPer_CAF1 124800 107 - 1 ---GUAUCUGACAGAUACAUUUG--------CUAGCGAUGCCUACAAACUGACAAAAAGUCAACUC--CAGCCUAGGCCCGGACGUGCCACAGUGCCGCCGCGUGGCGCAUUUGAAAAGA ---(((((.....))))).((((--------.(((......))))))).((((.....))))..((--(.((....))..))).(((((((.(((....))))))))))........... ( -28.40) >consensus AAAGUAUCUGACAGAUACAUAUG________CUCGUGCUGCCUACAAACUGACAAAAAGUCAACUU__CAGCCUAUGGCCAAACGUGCCGCCAU______GUGAGGCGCAUUUGAAA___ ...(((((.....)))))..................((((.........((((.....))))......)))).......((((.(((((...............))))).))))...... (-17.32 = -17.32 + 0.00)

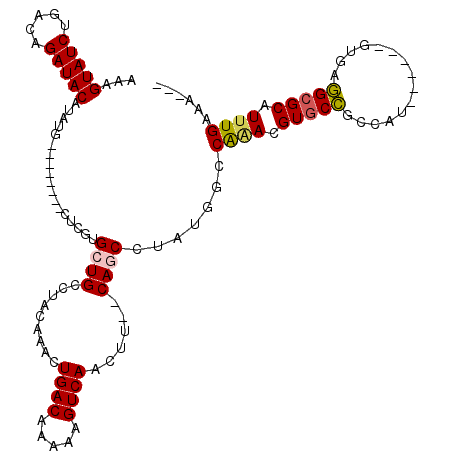

| Location | 4,972,546 – 4,972,656 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -29.86 |
| Energy contribution | -29.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4972546 110 - 27905053 GGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUGCCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCC (((((((((((((((((((((.....))).)))))))....(((((.....)))))))))((....))...........((((.....))))))))))).(((...))). ( -31.60) >DroSec_CAF1 79385 110 - 1 GGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUGCCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCC (((((((((((((((((((((.....))).)))))))....(((((.....)))))))))((....))...........((((.....))))))))))).(((...))). ( -31.60) >DroSim_CAF1 81197 110 - 1 GGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUGCCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCC (((((((((((((((((((((.....))).)))))))....(((((.....)))))))))((....))...........((((.....))))))))))).(((...))). ( -31.60) >DroEre_CAF1 80072 110 - 1 GGGAAGUACAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUGCCUACAAACUGACAAAGAGUCAUCUUUUCAGCCUAUAGCC (((.(((((..((((((((((.....))).)))))))....(((((.....)))))........))))).))).....((((.(((((....)))))))))......... ( -33.50) >DroYak_CAF1 81089 110 - 1 GGGAAGUACAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUGCCUACAAACUGACAAAGAGUCAACUUUUCAGCCUAUAGCC (((.(((((..((((((((((.....))).)))))))....(((((.....)))))........))))).))).....((((.((((......))))))))......... ( -31.30) >consensus GGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUGCCUACAAACUGACAAAGAGUCAACUUCUCAGCCUAUGGCC (((.(((((..((((((((((.....))).)))))))....(((((.....)))))........))))).)))......((((.....))))........((.....)). (-29.86 = -29.62 + -0.24)



| Location | 4,972,586 – 4,972,693 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 99.07 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -36.44 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4972586 107 - 27905053 AAACGUGCCCGGUGCAGUGAACACGGUUCGCUCUUCUGGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUG ..(((..(((((...(((((((...)))))))...))))).(((((((((((((((((.....))).)))))))....(((((.....))))))))))))))).... ( -38.70) >DroSec_CAF1 79425 107 - 1 AAACGUGCCCGGUGCAGUGAACACGGUUCGCUCUUCUGGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUG ..(((..(((((...(((((((...)))))))...))))).(((((((((((((((((.....))).)))))))....(((((.....))))))))))))))).... ( -38.70) >DroSim_CAF1 81237 107 - 1 AAACGUGCCCGGUGCAGUGAACACGGUUCGCUCUUCUGGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUG ..(((..(((((...(((((((...)))))))...))))).(((((((((((((((((.....))).)))))))....(((((.....))))))))))))))).... ( -38.70) >DroEre_CAF1 80112 107 - 1 AAACGUGCCCGGUGCAGUGAACACGGUUCGCUCUUCUGGGAAGUACAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUG .......(((((...(((((((...)))))))...))))).(((((..((((((((((.....))).)))))))....(((((.....)))))........))))). ( -38.30) >DroYak_CAF1 81129 107 - 1 AAACGUGCCCGGUGCAGUGAACACGGUUCGUUCUUCUGGGAAGUACAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUG .......(((((.....(((((...))))).....))))).(((((..((((((((((.....))).)))))))....(((((.....)))))........))))). ( -33.20) >consensus AAACGUGCCCGGUGCAGUGAACACGGUUCGCUCUUCUGGGAAGUAUAUGCCGGCUACGUGUAACGUGAGUUGGCCAAAGUAUCUGACAGAUACAUAUGCUCGUGCUG ..(((..(((((...(((((((...)))))))...))))).(((((((((((((((((.....))).)))))))....(((((.....))))))))))))))).... (-36.44 = -36.68 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:54 2006