| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,971,776 – 4,971,883 |
| Length | 107 |
| Max. P | 0.500000 |

| Location | 4,971,776 – 4,971,883 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.24 |
| Mean single sequence MFE | -18.02 |
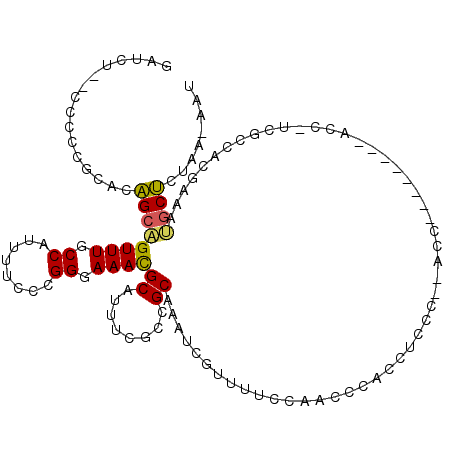
| Consensus MFE | -9.19 |
| Energy contribution | -8.88 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
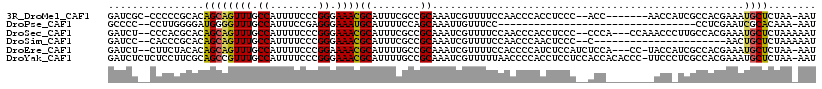
>3R_DroMel_CAF1 4971776 107 + 27905053 GAUCGC-CCCCCGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUCGCCGCAAAUCGUUUUCCAACCCACCUCCC--ACC-------AACCAUCGCCACGAAAUGCUCUAA-AAU ....((-.....))......((((.((........)).))))((((((((..((........................--...-------.......))..)))))))).....-... ( -17.28) >DroPse_CAF1 117394 82 + 1 GCCCC--CCUUGGGGAUGGGGUUUGCCAUUUCCGAGGGAAAUGCAUUUUCCAGCAAAUUGUUUCC---------------------------------CCUCGAAUCGCACAAA-AAU ((..(--((((((..((((......))))..)))))))...(((........)))..........---------------------------------.........)).....-... ( -25.70) >DroSec_CAF1 78624 111 + 1 GAUCU--CCCACGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUCGCCGCAAAUCGUUUUCCAACCCACCUCCC--CCCA---CCAAACCCUUGCCACGAAAUGCUCUAAAAAU .....--.............((((.((........)).))))((((((((..((((...((((...............--....---..))))..))))..))))))))......... ( -19.70) >DroSim_CAF1 80443 92 + 1 GAUCC--CACCCGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUCGCCGCAAAUCGUUUUCCAACCCAACUCCC--C----------------------AACUGCUCUAAAAAU .....--.........(((((((............((((((((.((((......))))))))))))............--.----------------------)))))))........ ( -16.71) >DroEre_CAF1 79315 111 + 1 GAUCU--CUUCUACACAGCAGUUUGCCAUUUUCCCGGAAAACGCAUUUUGCCGCAAAUCGUUUUCCACCCCAUCUCCAUCUCCA---CC-UACCAUCGCCACGAAAUGCUCUAA-AAU .....--.........((((...............((((((((.((((......))))))))))))..................---..-.....(((...)))..))))....-... ( -14.40) >DroYak_CAF1 80311 116 + 1 GAUCUCUCUCCUUCGCAGCCGUUUGCCAUUUUCCCGGGAAACGCAUUUUGCCGCAAAUCGUUUUUAACCCCACCUCCUCCACCACACCC-UUCCCUCGCCACGAAAUGCUCUAA-AAU ....................((((.((........)).))))((((((((..((...................................-.......))..)))))))).....-... ( -14.35) >consensus GAUCU__CCCCCGCACAGCAGUUUGCCAUUUUCCCGGGAAACGCAUUUCGCCGCAAAUCGUUUUCCAACCCACCUCCC__ACC________ACC_UCGCCACGAAAUGCUCUAA_AAU ................((((((((.((........)).))))((........))....................................................))))........ ( -9.19 = -8.88 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:51 2006