
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,968,610 – 4,968,710 |
| Length | 100 |
| Max. P | 0.645351 |

| Location | 4,968,610 – 4,968,710 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4968610 100 + 27905053 UCUCUUGGC-UUUUUGUUUAUAUUUUAAUUACGUUUUCGUUUUCGUAGUUGAUUUAUGCUCAUGAAAUGGCGUGGGUGGCAUUAAAAG-GGGGUGUGUG---GUA ((((((.((-(........(((..(((((((((..........)))))))))..)))(((((((......)))))))))).....)))-))).......---... ( -22.20) >DroPse_CAF1 113868 80 + 1 UCU----GC-UUUUUGUUUAUGUUUUAAUUACGUUU------UUGUGGUUGAUUUAUGCCCAUGAAAUGGCGUAGGCCGCACCGA-GUGGAG------------- ((.----.(-((..(((.......(((((((((...------.)))))))))((((((((........))))))))..)))..))-)..)).------------- ( -20.80) >DroSec_CAF1 75562 83 + 1 UCUCUUGGC-UUUUUGUUUAUAUUUUAAUUACGUUU------UCGUGGUUGAUUUAUGCUCAUGAAAUGGCGUGGGUGGCCUUAAAAAGA--------------- ......(((-(........(((..(((((((((...------.)))))))))..)))(((((((......))))))))))).........--------------- ( -21.30) >DroSim_CAF1 76569 98 + 1 UCUCUUGGC-UUUUUGUUUAUAUUUUAAUUACGUUU------UCGUGGUUGAUUUAUGCUCAUGAAAUGGCGUGGGUGGCCUUAAAAAGGGGGUGUGGGGUCGUA (((((((((-(........(((..(((((((((...------.)))))))))..)))(((((((......))))))))))).....))))))............. ( -24.00) >DroEre_CAF1 76188 96 + 1 UCUCUUGGCGUUUUUGUUUAUAUUCUAAUUACGUUU------CCGUGGUUGAUUUAUGCUCGAGAAAUGGCGUGGGUGGUAUUAAAAAGGGGGUGUGUG---GUA .(((.((.((((((((..((((...((((((((...------.))))))))...))))..).))))))).)).))).......................---... ( -16.20) >DroYak_CAF1 77157 95 + 1 UCUCUUGGCGUUUUUGUUUAUAUUUUAAUUACGUUU------UCGUGGUUGAUUUAUGCCCAUGAAACCGCGUGGGUGGUAUUAA-AAGGGGGUGUGUG---GAA ..(((..(((..((..((((((..(((((((((...------.)))))))))..)).(((((((......))))))).....)))-)..))..)))..)---)). ( -24.40) >consensus UCUCUUGGC_UUUUUGUUUAUAUUUUAAUUACGUUU______UCGUGGUUGAUUUAUGCUCAUGAAAUGGCGUGGGUGGCAUUAAAAAGGGGGUGUGUG___GUA ((((((.............(((..(((((((((..........)))))))))..)))(((((((......))))))).........))))))............. (-14.81 = -15.32 + 0.51)
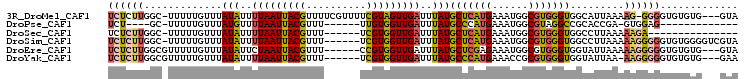
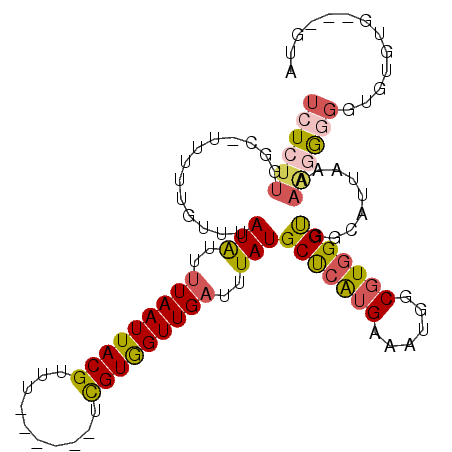
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:49 2006