| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
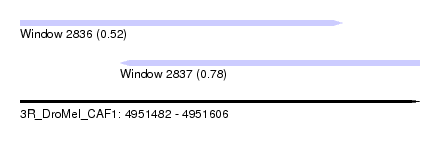
| Location | 4,951,482 – 4,951,606 |
| Length | 124 |
| Max. P | 0.784799 |

| Location | 4,951,482 – 4,951,582 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.87 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4951482 100 + 27905053 UAAAAUGCUUUGGCGAAGCGCAGCGAAAAUGCU--------AAUUAAUGUGAUUAUGUGGCCAUUUUAA-UGUUAAACAAUUGUUUAACGAUUAGUCAGGCGUGAG-AAC ....((((((((((...(((((.......)))(--------((((.....))))).)).))))..((((-((((((((....))))))).)))))..))))))...-... ( -20.10) >DroVir_CAF1 70967 98 + 1 GAA--ACC--AACUGAGGCGCAACGAAAAUGCU--------AAUUAAUGUGAUUAUGUUGCCAUUUAAAUUGUUAAACAAUUGUUUAGCGAUUAGUCAGGCGUCAAGAAU ...--...--...(((.((((((((.((.(((.--------.......))).)).))))))......(((((((((((....)))))))))))......)).)))..... ( -23.40) >DroPse_CAF1 94656 100 + 1 GAAGGCCCAACGGCAAGGCGCAACGAAAAUGCU--------AAUUAAUGUGAUUAUGUGGCCAUUUUAA-UGUUAAACAAUUGUUUAGCGAUUAGUCAGGCGUCAG-AGG ....(((....)))..(((((.((.((((((((--------(....(((....))).))).))))))..-((((((((....))))))))....))...)))))..-... ( -23.50) >DroGri_CAF1 68612 96 + 1 GAA--CGC--AUCUGAAGCGCAACGAAAUUGCU--------AAUUAAUGUGAUUAUGUUGCCAUUUGGA-UGCUAAACAAUUGUUUAGCGCUUAGUCAGGCGUCAU-AAG ((.--(((--..(((((((((((((.(((..(.--------.......)..))).))))))........-.(((((((....))))))))))...)))))))))..-... ( -31.20) >DroWil_CAF1 58781 100 + 1 AAUAAGGGCUAACCGAGGCGCAACGAAAAUGCU--------AAUUAAUGUGAUUAUGUGGCCAUUUUAA-UGUUAAACAAUUGUUUAACGAUUAGUCAGGCGUCAG-GGG ............((..(((((.((.((((((((--------(....(((....))).))).))))))..-((((((((....))))))))....))...))))).)-).. ( -22.00) >DroPer_CAF1 100727 108 + 1 GAAGGCCCAACGGCAAGGCGCAACGAAAAUGCUGCAACGAAAAUUAAUGUGAUUAUGUGGCCAUUUUAA-UGUUAAACAAUUGUUUAGCGAUUAGUCAGGCGUCAG-AGG ....(((....)))..(((((.((.(((((((..((......((....)).....))..).))))))..-((((((((....))))))))....))...)))))..-... ( -25.00) >consensus GAA__CCC__AGCCGAGGCGCAACGAAAAUGCU________AAUUAAUGUGAUUAUGUGGCCAUUUUAA_UGUUAAACAAUUGUUUAGCGAUUAGUCAGGCGUCAG_AAG ................((((((.......)))...........................(((...((((.((((((((....)))))))).))))...))))))...... (-16.70 = -16.87 + 0.17)



| Location | 4,951,513 – 4,951,606 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.63 |
| Mean single sequence MFE | -12.23 |
| Consensus MFE | -9.29 |
| Energy contribution | -9.45 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4951513 93 - 27905053 --UACCUACC----UAGCCAUCGCAUUUUCGUUCUCACGCCUGACUAAUCGUUAAACAAUUGUUUAACAU-UAAAAUGGCCACAUAAUCACAUUAAUUAG --........----..(((((........(((....)))...........(((((((....)))))))..-....))))).................... ( -12.40) >DroPse_CAF1 94687 81 - 1 UG------------------UGGGGCGGCUCCUCUGACGCCUGACUAAUCGCUAAACAAUUGUUUAACAU-UAAAAUGGCCACAUAAUCACAUUAAUUAG ((------------------(((((((.(......).)))).........(.(((((....))))).)..-........)))))................ ( -15.60) >DroSim_CAF1 58820 97 - 1 --UACCUACCCAUCUACCCAUCGUAUUUUCGUUCUCACGCCUGACUAAUCGUUAAACAAUUGUUUAACAU-UAAAAUGGCCACAUAAUCACAUUAAUUAG --....................................(((....((((.(((((((....)))))))))-))....))).................... ( -11.70) >DroEre_CAF1 59015 86 - 1 --C--A----------CCCAUCUUAUUUUUGUUCUCACGCCUGACUAAUCGUUAAACAAUUGUUUAACAUUUAAAAUGGCCACAUAAUCACAUUAAUUAG --.--.----------......................(((....(((..(((((((....)))))))..)))....))).................... ( -11.00) >DroWil_CAF1 58812 79 - 1 AG------------------UU--UCAUCUCCCCUGACGCCUGACUAAUCGUUAAACAAUUGUUUAACAU-UAAAAUGGCCACAUAAUCACAUUAAUUAG ..------------------..--..............(((....((((.(((((((....)))))))))-))....))).................... ( -11.70) >DroYak_CAF1 59889 88 - 1 --UGCC----------CCCGUCGUAUUUUUGUUCUCACGCCUGACUAAUCGUUAAACAAUUGUUUAACAUUUAAAAUGGCCACAUAAUCACAUUAAUUAG --....----------......................(((....(((..(((((((....)))))))..)))....))).................... ( -11.00) >consensus __U__C__________CCCAUCGUAUUUUUGUUCUCACGCCUGACUAAUCGUUAAACAAUUGUUUAACAU_UAAAAUGGCCACAUAAUCACAUUAAUUAG ......................................(((.........(((((((....))))))).........))).................... ( -9.29 = -9.45 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:46 2006