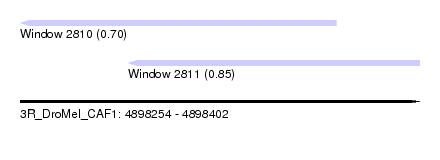
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,898,254 – 4,898,402 |
| Length | 148 |
| Max. P | 0.852728 |

| Location | 4,898,254 – 4,898,371 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -47.74 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.02 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4898254 117 - 27905053 CCGGCGACUGCAGCAGCUGCUCAGGCAGCGGUGGCCGCGGCUGCCAACAAUAUGUUCGGAUCCAACAGCUCGAUCUUCUCGAAUCCCCUGGCCAUUCCGGGUACCGCAGCUGUGGCA .........((.((((((((...((((((.((....)).)))))).......((..((((.....(((.((((.....)))).....))).....))))..))..)))))))).)). ( -46.80) >DroGri_CAF1 8707 117 - 1 CCGGCAACAGCAGCAGCCGCACAGGCGGCCGUUGCCGCGGCGGCCAACAAUAUGUUUGGCUCGAACAGUUCGCUCUUCUCGAGCCAUCUGGCCAUGCCGGGCACCACACCAGCAGCA ((((((...(((((.(((((....))))).)))))......(((((((.....)))((((((((..((......))..))))))))...)))).))))))((.........)).... ( -51.80) >DroWil_CAF1 7479 110 - 1 CCCACCACAGCGGCAGCUGCUCAGGCUGCUGUGGCAGCCGCUGCAAAUAACAUGUUCGGCUCGAAUAGUUCCCUUUUCUCCAAUCAUCUGGCCAUACCGGGAACCGCUG---U---- ......(((((((((((......((((((....)))))))))))........((((((...))))))((((((......(((......))).......)))))))))))---)---- ( -40.72) >DroYak_CAF1 6330 117 - 1 CCGGCGACUGCAGCAGCUGCCCAGGCAGCGGUGGCCGCGGCUGCCAACAAUAUGUUCGGAUCCAACAGCUCGAUCUUCUCGAACCCCCUGGCCAUUCCGGGUACCGCAGCCGUGGCA ((.((.......)).(((((....)))))))..(((((((((((.........(((((((((.........))))....)))))..(((((.....)))))....))))))))))). ( -49.80) >DroMoj_CAF1 7557 117 - 1 CCGGCCACAGCGGCAGCGGCCCAGGCGGCAGUUGCGGCCGCAGCGAACAAUAUGUUCGGGUCGAACAGUUCGCUCUUCUCGAAUCACUUGGCGCUGCCGGGCACAACGCCCGCCGCC .((((......(((((((.((.(((((((.......)))))(((((((....((((((...)))))))))))))............)).)))))))))((((.....)))))))).. ( -57.20) >DroAna_CAF1 1666 117 - 1 CCGGCAACAGCAGCAGCUGCCCAGGCAGCCGUGGCCGCCGCCGCCAAUAAUAUGUUCGGCUCGAAUAGCUCACUAUUUUCGAAUCACCUGGCCAUCCCGGGAACUGCAGCAGUGGCU ..(((..(.((.((.(((((....))))).)).)).)..)))((((......(((((((.((((((((....))))..))))....(((((.....)))))..))).)))).)))). ( -40.10) >consensus CCGGCAACAGCAGCAGCUGCCCAGGCAGCCGUGGCCGCCGCUGCCAACAAUAUGUUCGGCUCGAACAGCUCGCUCUUCUCGAAUCACCUGGCCAUGCCGGGAACCGCAGCAGUGGCA .........(((((.(((((....))))).(((((....)))))........((((((...)))))))))................(((((.....))))).............)). (-24.88 = -24.02 + -0.86)



| Location | 4,898,294 – 4,898,402 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 70.47 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4898294 108 - 27905053 GCAACAACAGCAGCUGGCCGCCGUUGGUCUGCCGGCGACUGCAGCAGCUGCUCAGGCAGCGGUGGCCGCGGCUGCCAACAAUAUGUUCGGAUCCAACAGCUCGAUCUU ((.....((((.((.(((((((....(.((((.(....).))))).(((((....)))))))))))))).))))(((((.....))).))........))........ ( -44.70) >DroPse_CAF1 33800 88 - 1 -----AACGACAGAUGG---CCGUGGGCCUGCCGGCCACCGCUGCGGCUGCCCAGG------------CGGCCGUCAACAACAUGUUCGGCUCAAACAGCUCGCUGUU -----.......(((((---((((((((..((((((....))..)))).))))..)------------))))))))...((((.((..((((.....)))).)))))) ( -36.90) >DroGri_CAF1 8747 95 - 1 ----------CAGCUGG---CUGUGGGCCUGCCGGCAACAGCAGCAGCCGCACAGGCGGCCGUUGCCGCGGCGGCCAACAAUAUGUUUGGCUCGAACAGUUCGCUCUU ----------.(((..(---((((.((((.((((....).(((((.(((((....))))).)))))...)))))))..(((.....)))......)))))..)))... ( -43.40) >DroWil_CAF1 7512 98 - 1 ----------CAAUUGGCUGCCGUUGGCUUGCCCACCACAGCGGCAGCUGCUCAGGCUGCUGUGGCAGCCGCUGCAAAUAACAUGUUCGGCUCGAAUAGUUCCCUUUU ----------.....(((((((((((............)))))))))))((...((((((....))))))...))....(((.((((((...)))))))))....... ( -39.10) >DroMoj_CAF1 7597 95 - 1 ----------CAGCUGG---CGGUCGGUCUGCCGGCCACAGCGGCAGCGGCCCAGGCGGCAGUUGCGGCCGCAGCGAACAAUAUGUUCGGGUCGAACAGUUCGCUCUU ----------..((((.---.((((((....)))))).))))....(((((((((.(....)))).))))))(((((((....((((((...)))))))))))))... ( -47.20) >consensus __________CAGCUGG___CCGUGGGCCUGCCGGCCACAGCAGCAGCUGCCCAGGCGGC_GUGGCCGCGGCUGCCAACAAUAUGUUCGGCUCGAACAGUUCGCUCUU ...........(((((.....(((.((....)).)))......((((((((................)))))))).....................)))))....... (-19.35 = -19.03 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:22 2006