
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,896,639 – 4,896,774 |
| Length | 135 |
| Max. P | 0.966492 |

| Location | 4,896,639 – 4,896,734 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -26.93 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4896639 95 + 27905053 ACUGCCUGG----CUUAAU-----UUGUUAUUUCCUUUACAGCACAACGUUGCCGUUGGUCGGGGGUCCAAUGGGACCCAAGCUGCUGUUGACAGCGCCUGACA .(((...((----..(((.-----...)))...))....)))..(((((....)))))(((((((((((....))))))..((.((((....))))))))))). ( -31.80) >DroPse_CAF1 32108 101 + 1 ACUACAUCGACCGUAUGAUUUCUUCUCUAAUUGUCUUUAUAGGACGCCGUUGGCGGCUGUGGGUGGUCCAAUGGGACCAAGGCCACUGUUGACGGCA---GCCA ........(((.((..((.....))....)).)))......((..(((((..(((...((((.((((((....))))))...)))))))..))))).---.)). ( -34.80) >DroSec_CAF1 4669 95 + 1 ACUGCCUGG----CUUAAU-----UUGUUAUUUCAUUUACAGCACAACGUUGCCGUUGGUCGGGGGUCCAAUGGGACCCAAUCCGCUGUUGACAGCGCCUGACA ........(----((....-----.((......)).....))).(((((....)))))(((((((((((....))))))....(((((....))))).))))). ( -32.80) >DroSim_CAF1 4601 95 + 1 ACUGCCUGG----CUUAAU-----UUGUUAUUUCCUUUACAGCACAACGUUGCCGUUGGUCGGGGGUCCAAUGGGACCCAAGCCGCUGUUGACAGCGCCUGACA .(((...((----..(((.-----...)))...))....)))..(((((....)))))(((((((((((....))))))....(((((....))))).))))). ( -31.90) >DroEre_CAF1 4215 94 + 1 ACUGCCUGG----CUUA------AUGGUUAUUUCAUUUACAGCACAACGUUGCCGUUGGUCGGGGGUCCAAUGGGACCCAAGCCGCUGUUGACAGCGCCUGACA ..(((.((.----...(------((((.....)))))..)))))(((((....)))))(((((((((((....))))))....(((((....))))).))))). ( -35.80) >DroYak_CAF1 4710 99 + 1 ACUGCCUGG----CUUAUU-UCCAUUACUAUUUGCUUUACAGCACAACGUUGCCGUUGGUCGGGGGUCCAAUGGGACCCAAGCCGCUGUUGACAGCGCCUGACA ...((..((----(((...-.((.........((((....))))(((((....)))))...))((((((....)))))))))))((((....))))))...... ( -35.10) >consensus ACUGCCUGG____CUUAAU_____UUGUUAUUUCCUUUACAGCACAACGUUGCCGUUGGUCGGGGGUCCAAUGGGACCCAAGCCGCUGUUGACAGCGCCUGACA ............................................(((((....)))))(((((((((((....))))))....(((((....))))).))))). (-26.93 = -27.02 + 0.09)



| Location | 4,896,639 – 4,896,734 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -26.17 |
| Energy contribution | -26.31 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4896639 95 - 27905053 UGUCAGGCGCUGUCAACAGCAGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAGGAAAUAACAA-----AUUAAG----CCAGGCAGU ((((.(((((((....))))(((..((((((....)))))).....(((((....))))).)))................-----.....)----)).)))).. ( -36.10) >DroPse_CAF1 32108 101 - 1 UGGC---UGCCGUCAACAGUGGCCUUGGUCCCAUUGGACCACCCACAGCCGCCAACGGCGUCCUAUAAAGACAAUUAGAGAAGAAAUCAUACGGUCGAUGUAGU .(((---.(((((.....(((((..((((((....))))))......)))))..))))))))(((((..(((.....((.......)).....)))..))))). ( -30.90) >DroSec_CAF1 4669 95 - 1 UGUCAGGCGCUGUCAACAGCGGAUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAUGAAAUAACAA-----AUUAAG----CCAGGCAGU ((((.(((((((....))))((...((((((....))))))))(..(((((....)))))..).................-----.....)----)).)))).. ( -35.10) >DroSim_CAF1 4601 95 - 1 UGUCAGGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAGGAAAUAACAA-----AUUAAG----CCAGGCAGU ((((....((((....))))(((((((((((....))))))((.(((((((....)))))....))...)).........-----...)))----)).)))).. ( -36.60) >DroEre_CAF1 4215 94 - 1 UGUCAGGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAUGAAAUAACCAU------UAAG----CCAGGCAGU ((((.(((.......(((((((...((((((....))))))))...(((((....))))).))))).((((.......)))------)..)----)).)))).. ( -36.50) >DroYak_CAF1 4710 99 - 1 UGUCAGGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAGCAAAUAGUAAUGGA-AAUAAG----CCAGGCAGU ((((....((((....))))(((((((((((....))))))(((..(((((....)))))((((....))))........))).-...)))----)).)))).. ( -41.40) >consensus UGUCAGGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAGGAAAUAACAA_____AUUAAG____CCAGGCAGU .(((((.(((((....))))).)).((((((....))))))..)))(((((....))))).(((((.................................))))) (-26.17 = -26.31 + 0.14)


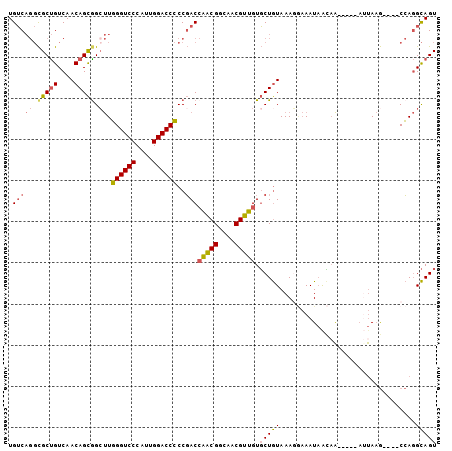
| Location | 4,896,657 – 4,896,774 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.03 |
| Mean single sequence MFE | -43.73 |
| Consensus MFE | -21.18 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4896657 117 - 27905053 CCCUCCUCCCCGGACGCCUCCGAGUCCUCGUCCUCGGUGGUGUCAGGCGCUGUCAACAGCAGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAGGAAAUAA ...((((..((.((((((.(((((........))))).)))))).))........((((((....((((((....)))))).....(((((....)))))))))))..))))..... ( -51.40) >DroPse_CAF1 32135 114 - 1 CCAACAUCGCCAGCUGUGUCCGAGUCGUCAUCGCCGGUGGUGGC---UGCCGUCAACAGUGGCCUUGGUCCCAUUGGACCACCCACAGCCGCCAACGGCGUCCUAUAAAGACAAUUA ...((((.(....).))))....(((.....(((((.(((((((---((..((((....))))..((((((....))))))....))))))))).))))).........)))..... ( -42.24) >DroWil_CAF1 5805 114 - 1 CCCACAUCCCCAGCCCUUUCCGAAUCAUCCUCAUCUGUGGUGACG---GCCAUCAAUGCUGGCCUAGGUCCAAUUGGACCACCCACAACAGCCAAUGGAGUGCUGUAAAUACAACAA ..........((((...((((((.......))..((((.(((..(---((((.......)))))..(((((....)))))...))).)))).....)))).))))............ ( -32.40) >DroYak_CAF1 4732 117 - 1 CCCUCCUCCCCGGACGCCUCCGAGUCCUCGUCGUCGGUGGUGUCAGGCGCUGUCAACAGCGGCUUGGGUCCCAUUGGACCCCCGACCAACGGCAACGUUGUGCUGUAAAGCAAAUAG ..........((((((((.(((((.......).)))).))))))((.(((((....))))).)).((((((....)))))).))..(((((....)))))((((....))))..... ( -48.80) >DroAna_CAF1 23 114 - 1 CCCUCCUCCCCAUCCGCCUCGGAGUCCUCAUCCUCGGUGGUUUCGGCAGCGGUGAACAGUGGCUUGGGUCCCAUCGGGCCGCCCACCACCGCCAAUGGGGUGCUGUAAG---AAUAU .....(.(((((((((((..(((.......)))..)))))....(((.(.((((....(((((((((......))))))))).)))).).))).)))))).).......---..... ( -44.70) >DroPer_CAF1 38783 114 - 1 CCAACAUCGCCAGCUGUGUCCGAGUCGUCAUCGCCGGUGGUGGC---UGCCGUCAACAGUGGCCUUGGUCCCAUUGGACCACCCACAGCCGCCAACGGCGUCCUAUAAAGACAAGUA ............(((.((((...........(((((.(((((((---((..((((....))))..((((((....))))))....))))))))).))))).........))))))). ( -42.85) >consensus CCCACAUCCCCAGCCGCCUCCGAGUCCUCAUCGUCGGUGGUGUC___CGCCGUCAACAGUGGCCUGGGUCCCAUUGGACCACCCACAACCGCCAACGGCGUGCUGUAAAGACAAUAA ...............................(((((.((((((.....((((.......))))..((((((....)))))).......)))))).)))))................. (-21.18 = -21.85 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:20 2006