| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 826,168 – 826,281 |
| Length | 113 |
| Max. P | 0.908248 |

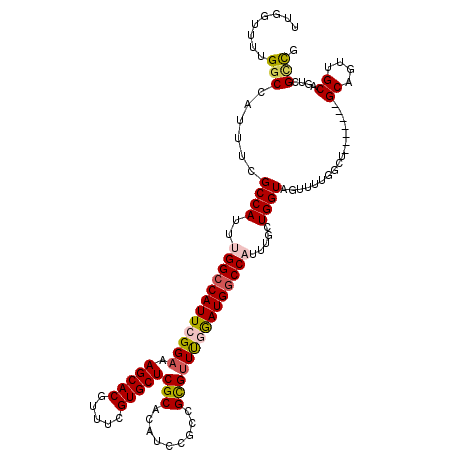
| Location | 826,168 – 826,281 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -24.92 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 826168 113 + 27905053 CGGCGAUUGCGACUGC-------AGCCAAAACUACCAGCUAAUGGCCAUCGAAACACGGCGGAUGUGCGAGCACGAAACGUGCUUUCCGAAUGGCCAAAUGGCGAAAUGGUCAAAACCAA .(((...(((....))-------))))..........((((.(((((((((...((((.....)))).(((((((...)))))))..)).)))))))..))))....((((....)))). ( -36.00) >DroSec_CAF1 8443 120 + 1 CGGCGACUGCAACUGCAGCCUGCAGCCAAAACUACCAGCAAUGGGCCAUCCAAACGCGGCGGAUGUGCGAGCACGAAACGUGCUUUCCGAAUGGCCAAAUGGCGAAAUGGCCAAAACCAA .((((.((((....)))).)....((((.......((....))((((((((......))((((.....(((((((...))))))))))).))))))...))))......)))........ ( -40.40) >DroSim_CAF1 5638 120 + 1 CGGCGACUGCAACUGCAGCCUGCAGCCAAAACUACCAGCAAUGGGCCAUCCAAACGCGGCGGAUGUGCGAGCACGAAACGUGCUUUCCGAAUGGCCAAAUGGCGAAAUGGCCAAAACCAA .((((.((((....)))).)....((((.......((....))((((((((......))((((.....(((((((...))))))))))).))))))...))))......)))........ ( -40.40) >DroEre_CAF1 10141 95 + 1 CGGC-------------------------AACUACCAACAAAUGGUCAUUCAAACGCGGCGGAUGUGCGAGCACGAAACGUGCUUUCCGAAUGGCCAAAUGGCGAAAUUGCCAAAACCAU .(((-------------------------((.......((..(((((((((...((((.......))))((((((...))))))....)))))))))..))......)))))........ ( -31.12) >DroYak_CAF1 8901 112 + 1 CAACGACUGCAACCGC-------AUCCAAAACUACCAACAAAUGGCCAUCUGAGCGCGGCGGAUGUGCGAGCACUAAACGUGCUUUCCGAAUGGCCAAAUGGCGAAAUUGCCAAAACC-C ....((.(((....))-------)))................((((((((.(((((((.....)))))((((((.....)))))))).).)))))))..(((((....))))).....-. ( -32.80) >consensus CGGCGACUGCAACUGC_______AGCCAAAACUACCAGCAAAUGGCCAUCCAAACGCGGCGGAUGUGCGAGCACGAAACGUGCUUUCCGAAUGGCCAAAUGGCGAAAUGGCCAAAACCAA ...........................................((((((((......))((((.....((((((.....)))))))))).))))))...((((......))))....... (-24.92 = -24.52 + -0.40)

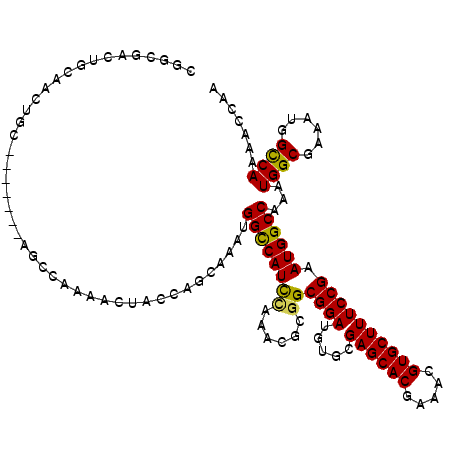
| Location | 826,168 – 826,281 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 826168 113 - 27905053 UUGGUUUUGACCAUUUCGCCAUUUGGCCAUUCGGAAAGCACGUUUCGUGCUCGCACAUCCGCCGUGUUUCGAUGGCCAUUAGCUGGUAGUUUUGGCU-------GCAGUCGCAAUCGCCG .((((....))))....((....(((((((.((((.((((((...)))))).((((.......)))))))))))))))...)).((..(((.((((.-------...)))).)))..)). ( -37.30) >DroSec_CAF1 8443 120 - 1 UUGGUUUUGGCCAUUUCGCCAUUUGGCCAUUCGGAAAGCACGUUUCGUGCUCGCACAUCCGCCGCGUUUGGAUGGCCCAUUGCUGGUAGUUUUGGCUGCAGGCUGCAGUUGCAGUCGCCG ..(((...(((((..(..(((...(((((((((((.((((((...))))))(((.........))))))))))))))......)))..)...)))))...((((((....))))))))). ( -46.90) >DroSim_CAF1 5638 120 - 1 UUGGUUUUGGCCAUUUCGCCAUUUGGCCAUUCGGAAAGCACGUUUCGUGCUCGCACAUCCGCCGCGUUUGGAUGGCCCAUUGCUGGUAGUUUUGGCUGCAGGCUGCAGUUGCAGUCGCCG ..(((...(((((..(..(((...(((((((((((.((((((...))))))(((.........))))))))))))))......)))..)...)))))...((((((....))))))))). ( -46.90) >DroEre_CAF1 10141 95 - 1 AUGGUUUUGGCAAUUUCGCCAUUUGGCCAUUCGGAAAGCACGUUUCGUGCUCGCACAUCCGCCGCGUUUGAAUGACCAUUUGUUGGUAGUU-------------------------GCCG ........((((((((((.((..(((.((((((((.((((((...))))))(((.........))))))))))).)))..)).))).))))-------------------------))). ( -32.80) >DroYak_CAF1 8901 112 - 1 G-GGUUUUGGCAAUUUCGCCAUUUGGCCAUUCGGAAAGCACGUUUAGUGCUCGCACAUCCGCCGCGCUCAGAUGGCCAUUUGUUGGUAGUUUUGGAU-------GCGGUUGCAGUCGUUG .-((((.((((......))))...))))...(((..(((((.....))))).(((.....((((((..((((..((((.....))))...))))..)-------))))))))..)))... ( -38.90) >consensus UUGGUUUUGGCCAUUUCGCCAUUUGGCCAUUCGGAAAGCACGUUUCGUGCUCGCACAUCCGCCGCGUUUGGAUGGCCAUUUGCUGGUAGUUUUGGCU_______GCAGUUGCAGUCGCCG ........(((......((((..((((((((((((.(((((.....)))))(((.........))))))))))))))).....)))).................((....))....))). (-27.12 = -27.86 + 0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:44 2006