
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,872,078 – 4,872,250 |
| Length | 172 |
| Max. P | 0.823402 |

| Location | 4,872,078 – 4,872,179 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -6.93 |
| Energy contribution | -8.45 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
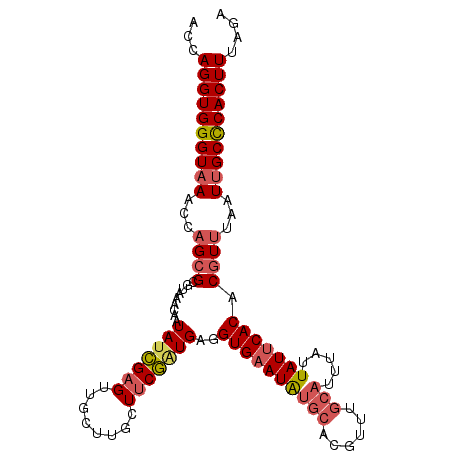
>3R_DroMel_CAF1 4872078 101 - 27905053 AUUUAUUAUUCACACGUUUAAUUGCUCACUUUAGAAUGCUUUAGUCAAUUAAUUAUAGGUUGCAUUUGCUACCUUUUAAUCUUUUUAUUUAAGCAGUCUGU-------------- ...............................((((.(((((.(((.((...((((.((((.((....)).))))..))))...)).))).))))).)))).-------------- ( -16.00) >DroSec_CAF1 3658 101 - 1 AUUUAUUAUUCACACGUUUAAUUGCCCACUUUAGACUGCUUUAGUCAUUUAAUUAUAGGUUCCAUUUGCUGCCUUUUAAUCUUUUUGGUUAAGCAGUCUGU-------------- ...............................((((((((((.(..((....((((.((((..(....)..))))..)))).....))..))))))))))).-------------- ( -20.40) >DroSim_CAF1 3694 101 - 1 AUUUAUUAUUCACACGUUUAAUUGCCCACUUUAGACUGCUUUAGUCAUUUAAUUAUAGGUUCUAUUUGCUACCUUUUAAUUAUUUUGGUUAAGCAGUCUGU-------------- ...............................((((((((((.(..((..((((((.((((..........))))..))))))...))..))))))))))).-------------- ( -22.70) >DroEre_CAF1 4595 92 - 1 AUAUAUUAUUCACACGUUUAAUAGCUCACUUUAGGCUGCUUAAUUCAUUUAAUUAUAGGUUUCAUUUGACACCUUUUAGUCUCUUUGGCCAA----------------------- ...(((((..(....)..)))))((.((....((((((..(((((.....))))).((((.((....)).))))..))))))...))))...----------------------- ( -11.00) >DroYak_CAF1 3742 115 - 1 AUUUGUUAUUCACACGUUUAAUUGCUCACUUGAGGCUGCUUAAUUCAUUUAAUUAUAGCUUCCAUUUGACACCUUUUAGUCUCUUUGGCUAAACGGGCUGUUAACUGUAAACUAU ...............(((((...........(((((((..(((((.....))))))))))))((.((((((((((((((((.....))))))).))).)))))).)))))))... ( -24.90) >consensus AUUUAUUAUUCACACGUUUAAUUGCUCACUUUAGACUGCUUUAGUCAUUUAAUUAUAGGUUCCAUUUGCUACCUUUUAAUCUUUUUGGUUAAGCAGUCUGU______________ ................................(((((((((..........((((.((((..(....)..))))..))))..........)))))))))................ ( -6.93 = -8.45 + 1.52)



| Location | 4,872,144 – 4,872,250 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.95 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -20.96 |
| Energy contribution | -22.27 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4872144 106 - 27905053 ACCAGGUGGGUAAACCAGCGCGUAAACAAUAUCGAGUUGCUUGCUUCGAUGAGGUGAAUAUGCACGUUUGCAUUUAUUAUUCACACGUUUAAUUGCUCACUUUAGA ...((((((((((...((((.........((((((((.....)).))))))..(((((((((((....)))).....))))))).))))...)))))))))).... ( -28.10) >DroSec_CAF1 3724 106 - 1 ACCAGGUGGGUAAACCUGCGCGUAAACAAUAUCGAGUUGCUUGUUUCGGUGAGGUGAAUAUGCACGUUUGCAUUUAUUAUUCACACGUUUAAUUGCCCACUUUAGA ...((((((((((...((.((((......(((((((........)))))))..(((((((((((....)))).....))))))))))).)).)))))))))).... ( -30.40) >DroSim_CAF1 3760 106 - 1 ACCAGGUGGGUAAACCAGCGCAUAAACAAUAUCGAGUUGCUUGCUUCGGUGAGGUGAAUAUGCACGUUUGCAUUUAUUAUUCACACGUUUAAUUGCCCACUUUAGA ...((((((((((...((((...................((..(....)..))(((((((((((....)))).....))))))).))))...)))))))))).... ( -31.90) >DroEre_CAF1 4652 101 - 1 ACCAGGUGGGUAAAAAAGGGUG-AAACAAUACUGUGUUGCUUGCUCCAAUGAAGUGUAUG----UGUUUACAUAUAUUAUUCACACGUUUAAUAGCUCACUUUAGG .((((((((((......(((..-(((((......))))..)..).))..((((..(((((----((....)))))))..))))...........))))))))..)) ( -22.20) >consensus ACCAGGUGGGUAAACCAGCGCGUAAACAAUAUCGAGUUGCUUGCUUCGAUGAGGUGAAUAUGCACGUUUGCAUUUAUUAUUCACACGUUUAAUUGCCCACUUUAGA ...((((((((((...((((.........(((((((........)))))))..((((((((((......))).....))))))).))))...)))))))))).... (-20.96 = -22.27 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:09 2006