| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,860,669 – 4,860,762 |
| Length | 93 |
| Max. P | 0.931951 |

| Location | 4,860,669 – 4,860,762 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -21.15 |
| Energy contribution | -20.32 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
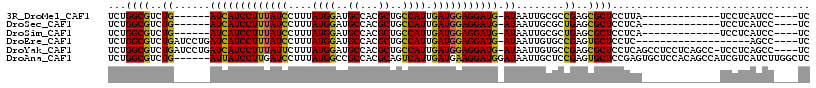
>3R_DroMel_CAF1 4860669 93 + 27905053 UCUGGCGUCUG------AUCAUCCUUUAUCCUUUAUGGAUGCCACGCUGCCAUUGAUGGAGGAUG-AUAAUUGCGCCGAGCGCUCCUUA-------------UCCUCAUCC----UC ...(((((...------(((((((((((((....((((..((...))..)))).)))))))))))-))....)))))(((.(.......-------------.))))....----.. ( -31.50) >DroSec_CAF1 41151 93 + 1 UCUGGCGUCUG------AUCAUCCUUUAUCCUUUAUGGAUGCCACGCUGCCAUUGAUGGAGGAUG-AUAAUUGCGCUGAGCGCUCCUCA-------------UCCUCAUCC----UC ..((((((.((------..(((((............))))).)).)).))))..((((.((((((-(.....((((...))))...)))-------------)))))))).----.. ( -31.10) >DroSim_CAF1 43167 93 + 1 UCUGGCGUCUG------AUCAUCCUUUAUCCUUUAUGGAUGCCACGCUGCCAUUGAUGGAGGAUG-AUAAUUGCGCUGAGCGCUCCUCA-------------UCCUCAUCC----UC ..((((((.((------..(((((............))))).)).)).))))..((((.((((((-(.....((((...))))...)))-------------)))))))).----.. ( -31.10) >DroEre_CAF1 41524 93 + 1 UCUGGCGUCUGAUCCUGAUCAUCCUUUAUCCUUUAUGGAUGCCACGCUGCCAUUGAUGGAGGAUG-AUAAUUGUGCCGAGUGCUCCUC-------------------AGCC----UC ...((((.(.(((....(((((((((((((....((((..((...))..)))).)))))))))))-)).))))))))(((.(((....-------------------))))----)) ( -29.50) >DroYak_CAF1 41733 111 + 1 UCUGGCGUCUGAUCCUGAUCAUCCUUUAUUCUUUAUGGAUGCCACGCUGCCAUUGAUGGAGGAUG-AUAAUUGUGCCGAGCGCUCCUCAGCCUCCUCAGCC-UCCUCAGCC----UC ...(((....((..((((((((((((((((....((((..((...))..)))).)))))))))))-)..........(((.(((....)))))).))))..-))....)))----.. ( -32.60) >DroAna_CAF1 42570 111 + 1 UCUGGCGUCUG------AUUAUCCUUGAUCCUUUAUGGCCGCCACGCAGUCAUUGAUGAAGGAUGGAUAAUUGCUCCGAGUGCUCCGAGUGCUCCACAGCCAUCGUCAUCUUGGCUC ...((.((..(------(((((((...((((((((((((.((...)).)))))....))))))))))))))))).))(((..(.....)..)))...(((((.........))))). ( -34.50) >consensus UCUGGCGUCUG______AUCAUCCUUUAUCCUUUAUGGAUGCCACGCUGCCAUUGAUGGAGGAUG_AUAAUUGCGCCGAGCGCUCCUCA_____________UCCUCAUCC____UC ...((((..((......(((((((((((((....((((..((...))..)))).))))))))))).))........))..))))................................. (-21.15 = -20.32 + -0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:00 2006