| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,825,005 – 4,825,116 |
| Length | 111 |
| Max. P | 0.922298 |

| Location | 4,825,005 – 4,825,116 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -26.23 |
| Energy contribution | -26.86 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
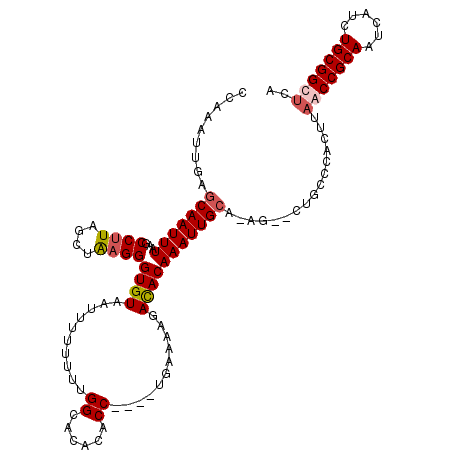
>3R_DroMel_CAF1 4825005 111 + 27905053 UGAGCCGCAGAUGAUUGCGGCUAAGUGGGCAAGGCUGUGCAAUUUGUGUCUUUACA----GGUGUGUGCCAAAAAAAAUUACACCCUUAGCUAAGGCUUAAAUUGCUCAAUUUGG ..((((((((....)))))))).......((((..((.((((((((.(((((...(----((.(((((...........)))))))).....))))).)))))))).)).)))). ( -35.70) >DroSec_CAF1 5879 115 + 1 UGAGCCGCAGAUGAUUGCGGCUAAGUGGGCAGGGCUGUGCAAUUUGUGUCUUUUCAGCUAGGUGUGUGCCAAAAAAAAUUACACCCUUAGCUAAGGCUUAAAUUGUUCAAUUUGG ..((((((((....)))))))).......((((..((.((((((((.(((((...(((((((.(((((...........))))).)))))))))))).)))))))).)).)))). ( -39.00) >DroEre_CAF1 5956 104 + 1 UGAGCCGCAGAUGAUUGCGGUUAAGUGGGAAG--CU-UGCAAUUUGUAUCUUUUUA----GGUGUGUGCCAAAAAAAAUUACACCCUCAGCUAAGGCUAAAAUUGCUCAAU---- ..((((((((....))))))))((((.....)--))-)(((((((...........----((((((.............))))))...(((....))).))))))).....---- ( -25.52) >DroYak_CAF1 5930 91 + 1 UGAGCCGCAGAUGAUUGCGGU----------------UGCAAUUUGUAUCUUUUCA----GGUGUGUGCCAA----AAUUACACCCUUAGCUAGGGCUAAAAUUGCUCAAUUUGG ..((((((((....)))))))----------------)(((((((..........(----((.(((((....----...))))))))((((....)))))))))))......... ( -26.20) >consensus UGAGCCGCAGAUGAUUGCGGCUAAGUGGGCAG__CU_UGCAAUUUGUAUCUUUUCA____GGUGUGUGCCAAAAAAAAUUACACCCUUAGCUAAGGCUAAAAUUGCUCAAUUUGG ..((((((((....))))))))................((((((((.(((((........((((((.............)))))).......))))).))))))))......... (-26.23 = -26.86 + 0.63)



| Location | 4,825,005 – 4,825,116 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -18.39 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4825005 111 - 27905053 CCAAAUUGAGCAAUUUAAGCCUUAGCUAAGGGUGUAAUUUUUUUUGGCACACACC----UGUAAAGACACAAAUUGCACAGCCUUGCCCACUUAGCCGCAAUCAUCUGCGGCUCA ......((.((((......((((....))))(((((((((.((((.(((......----))).))))...)))))))))....)))).))...(((((((......))))))).. ( -28.00) >DroSec_CAF1 5879 115 - 1 CCAAAUUGAACAAUUUAAGCCUUAGCUAAGGGUGUAAUUUUUUUUGGCACACACCUAGCUGAAAAGACACAAAUUGCACAGCCCUGCCCACUUAGCCGCAAUCAUCUGCGGCUCA .................(((....))).((((((((((((((((..((.........))..)))))).....)))))...)))))........(((((((......))))))).. ( -28.10) >DroEre_CAF1 5956 104 - 1 ----AUUGAGCAAUUUUAGCCUUAGCUGAGGGUGUAAUUUUUUUUGGCACACACC----UAAAAAGAUACAAAUUGCA-AG--CUUCCCACUUAACCGCAAUCAUCUGCGGCUCA ----..(((((....(((((....)))))((((((((((((((((((.......)----)))))))).....))))))-..--...))).......((((......))))))))) ( -24.70) >DroYak_CAF1 5930 91 - 1 CCAAAUUGAGCAAUUUUAGCCCUAGCUAAGGGUGUAAUU----UUGGCACACACC----UGAAAAGAUACAAAUUGCA----------------ACCGCAAUCAUCUGCGGCUCA .........(((((((..(((((.....)))))(((.((----((..((......----))..)))))))))))))).----------------.(((((......))))).... ( -22.80) >consensus CCAAAUUGAGCAAUUUAAGCCUUAGCUAAGGGUGUAAUUUUUUUUGGCACACACC____UGAAAAGACACAAAUUGCA_AG__CUGCCCACUUAACCGCAAUCAUCUGCGGCUCA .........(((((((...((((....))))((((..........((......))...........)))))))))))................(((((((......))))))).. (-18.39 = -19.20 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:49 2006