
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,806,417 – 4,806,564 |
| Length | 147 |
| Max. P | 0.956494 |

| Location | 4,806,417 – 4,806,536 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -20.23 |
| Energy contribution | -21.68 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4806417 119 - 27905053 CUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGAGGAGACAUUAAGGGAA-AAUGUAAGAGAGCAGUAUUGGGGAUAGUUUAAAGGUGCUAAUUAUUUCCAUUUAUAAAUA ........(.((((((.((((((....))))))..(((((((((....).)))))))).-......))))))).(((.(((..((((((..........))))))..)))..)))..... ( -40.30) >DroSec_CAF1 41824 120 - 1 CUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGGAGAGACAUUAAGGGAAAAAAGUAAGAGAGCAGUAUUGGGGAUAGUUUCAAGGUGCUAAUUAUUUCCAUUUAUAAAUA (((((.(((((((((..((((((.(.......).))))))..))))))).((....)).........)).)))))...(((..((((((..........))))))..))).......... ( -38.50) >DroSim_CAF1 38009 120 - 1 CUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGGAGAGAAAUUAAGGGAAAAAAGUAAGAGAGCAGUAUUGGGGAUAGUUUCAAGGUGCUAAUUAUUUCCAUUUAUAAAUA (((((.((.(((((...((((((....)))))).(((((...)))))......))))).........)).)))))...(((..((((((..........))))))..))).......... ( -36.70) >DroEre_CAF1 42843 117 - 1 CUGUUGUUAUCUCUUUG--GGAGCGUGCUCCCUGCGCUCUUGGGGAGACAUUAAGAGAA-AUAAUACGAGAGCAAUAUUGGGGAUAGUUUCAAGGUGCUAAUUAUUUCCAUUUAUAAAUA .........((((((..--(((((((.......)))))))..))))))......(.(((-(((((.....((((...(((..(....)..)))..)))).)))))))))........... ( -35.40) >DroYak_CAF1 43989 117 - 1 CUGUUGUUGUCUCUCUG--GGAGCGUGCUCCCCACUCUCUUGGGGAGACAUUAAGAGAA-AUAAUACGAGAGCUAUUUGGGGGAUAGUUUCAAGGUGCUAAUUAUUUUCAUCUAUAAAUA ((.(((.((((((((..--((((.(((.....))).))))..)))))))).))).))((-(((((.....(((..((((..(.....)..))))..))).)))))))............. ( -35.50) >DroAna_CAF1 42481 103 - 1 CUGUUGUUGGCUCUCUGAGAAAG------CUCCACUCUCUUGGGGGG-CAUAAAACUAA-AGAGAACGACAGCUCAAGGGUGGCCC---AUGGGGAGCUAAUAAAAAACAUUUU------ ...((((((((((((((((....------)))..(((....)))(((-(.....(((..-.(((........)))...))).))))---..)))))))))))))..........------ ( -31.70) >consensus CUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGGGGAGACAUUAAGGGAA_AAAGUAAGAGAGCAGUAUUGGGGAUAGUUUCAAGGUGCUAAUUAUUUCCAUUUAUAAAUA ..(((.(((((((((((((((((.(.......).))))))))))))))).((....)).........)).)))......((((((((((..........))))))))))........... (-20.23 = -21.68 + 1.45)


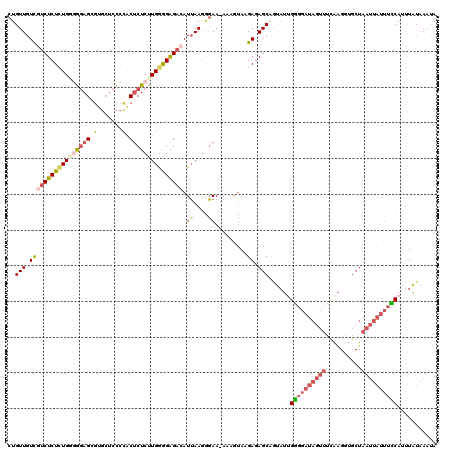
| Location | 4,806,457 – 4,806,564 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -19.22 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4806457 107 - 27905053 CAAAACACGCGAUGU------------AUUUAGCGAGCAGCUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGAGGAGACAUUAAGGGAA-AAUGUAAGAGAGCAGUAUUG (((.((..((((((.------------...((((.....)))).))))))((((((.((((((....))))))..(((((((((....).)))))))).-......))))))..)).))) ( -39.10) >DroSec_CAF1 41864 108 - 1 CAAAACACGCGAUGU------------AUUUAGCGAGCAGCUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGGAGAGACAUUAAGGGAAAAAAGUAAGAGAGCAGUAUUG .......(((.....------------.....)))....((((((.(((((((((..((((((.(.......).))))))..))))))).((....)).........)).)))))).... ( -35.20) >DroSim_CAF1 38049 108 - 1 CAAAACACGCGAUGU------------AUUUAGCGAGCAGCUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGGAGAGAAAUUAAGGGAAAAAAGUAAGAGAGCAGUAUUG (((.((..((((((.------------...((((.....)))).))))))((((((.((((((....)))))).(((((...)))))...................))))))..)).))) ( -34.20) >DroEre_CAF1 42883 105 - 1 CAAAACACGCGAUGU------------AUUUGGCGAGCAGCUGUUGUUAUCUCUUUG--GGAGCGUGCUCCCUGCGCUCUUGGGGAGACAUUAAGAGAA-AUAAUACGAGAGCAAUAUUG .......(((.(...------------...).)))....(((.((((..((((((..--(((((((.......)))))))..)))))).((((......-.)))))))).)))....... ( -32.60) >DroYak_CAF1 44029 105 - 1 CAAAACACGCGAUGU------------AUUUGGCGAGCAGCUGUUGUUGUCUCUCUG--GGAGCGUGCUCCCCACUCUCUUGGGGAGACAUUAAGAGAA-AUAAUACGAGAGCUAUUUGG ((((...(((.(...------------...).)))...((((.((((((((((((..--((((.(((.....))).))))..)))))))).........-.....)))).)))).)))). ( -36.73) >DroAna_CAF1 42512 112 - 1 CAAAACAUGCCAUCAGUUGGGGCCCCAACUUGGGGAGCAGCUGUUGUUGGCUCUCUGAGAAAG------CUCCACUCUCUUGGGGGG-CAUAAAACUAA-AGAGAACGACAGCUCAAGGG .......((((..(((((((....))))).))..).)))(((((((((.((((((..(((.((------.....)).)))..)))))-)......((..-..)))))))))))....... ( -40.30) >consensus CAAAACACGCGAUGU____________AUUUAGCGAGCAGCUGUUGUCGUCUCUCUGGGGGAGCGUGCUCCCCACUCUCUUGGGGAGACAUUAAGGGAA_AAAGUAAGAGAGCAGUAUUG ........((....................((((.....)))).....(((((((((((((((.(.......).)))))))))))))))......................))....... (-19.22 = -20.17 + 0.95)

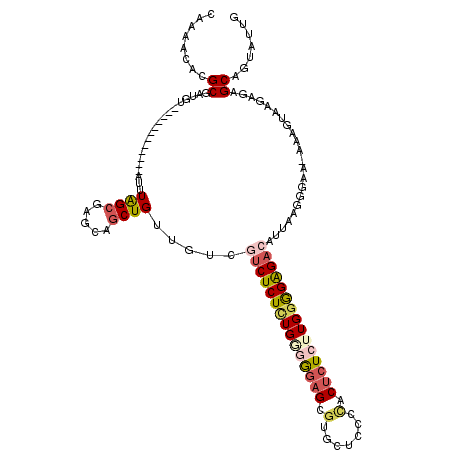
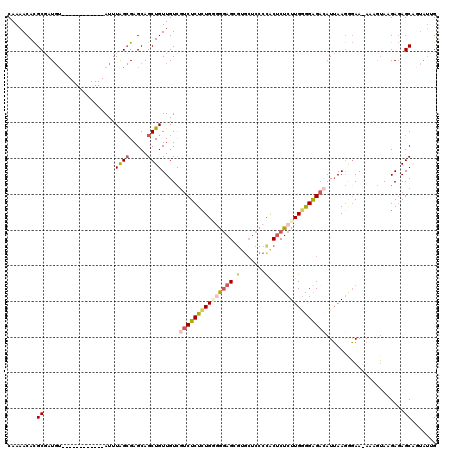
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:42 2006