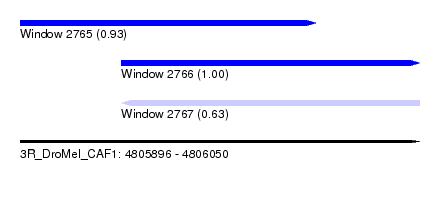
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,805,896 – 4,806,050 |
| Length | 154 |
| Max. P | 0.998551 |

| Location | 4,805,896 – 4,806,010 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -18.35 |
| Consensus MFE | -13.60 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4805896 114 + 27905053 GGACAAACAAUUUU-UUUUCCCGCUUCUUUAAAAUUGAGUAGCUA----UGUACAGAAAUAAAACAAAUUAAACUGAUCUUGACUUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUC ((((..........-.......(((.(((.......))).)))..----........................((((...((((((((........))))))))))))...)))).... ( -22.60) >DroSec_CAF1 41213 94 + 1 GGACAAACAAUUUUCUCUUUCCGCUUAUUUAAA-------------------------AUAAAACAUAUUAAACUGAUCUUGACAUCCAGAGACCAGGAAGUCAUCAGAAAGUUCCAUC ((((............................(-------------------------(((.....))))...((((...((((.(((........))).))))))))...)))).... ( -12.80) >DroSim_CAF1 37427 94 + 1 GGACAAUCAAUCUACUCUUUCCGCUUAUUUAAA-------------------------AUAAAACAAAUUAAACUGAGCUUGACAUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUC (((.............(((((.((((((((((.-------------------------..........))))).))))).((((.(((........))).))))...))))).)))... ( -16.04) >DroEre_CAF1 42207 112 + 1 -GACAGUAAACUUGAUAUUUCCGCUUAUUU--AAUUAAAUAACUA----UAUAAAGGAAUAAAACAAAUUAAACUGACCUUGACUUCCCCAGAUCAGGAAGUCAUCAGAAAGUUCCAUC -.......(((((.....((((..((((.(--(.((....)).))----.)))).))))..............((((...((((((((........)))))))))))).)))))..... ( -19.40) >DroYak_CAF1 43349 119 + 1 AGACAGUAAACUUUAUAUUUCCGCUUAGGUAAAAUUAAAUAACUACAUAUAUUAAGGAAUAAAACAUAUUGAACUGACAUUGACUUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUC ........((((((....((((...((.(((............)))...))....))))..............((((...((((((((........))))))))))))))))))..... ( -20.90) >consensus GGACAAUCAAUUUUAUAUUUCCGCUUAUUUAAAAUU_A_UA_CUA____U_U__AG_AAUAAAACAAAUUAAACUGACCUUGACUUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUC .........................................................................((((...((((((((........))))))))))))........... (-13.60 = -14.00 + 0.40)


| Location | 4,805,935 – 4,806,050 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4805935 115 + 27905053 AGCUA----UGUACAGAAAUAAAACAAAUUAAACUGAUCUUGACUUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUCGAAAUAUUGAUUAGGAAAGGCUACAGCAAAAUAAAUACAA ((((.----........................((((...((((((((........))))))))))))....(((((((((....)))))..)))).)))).................. ( -23.70) >DroSec_CAF1 41246 101 + 1 ------------------AUAAAACAUAUUAAACUGAUCUUGACAUCCAGAGACCAGGAAGUCAUCAGAAAGUUCCAUCGAAAUAUUGAUUAGGAAAGGCAACAGCAAAAUAAAUACAA ------------------...............((((...((((.(((........))).))))))))....(((((((((....)))))..)))).(....)................ ( -17.10) >DroSim_CAF1 37460 101 + 1 ------------------AUAAAACAAAUUAAACUGAGCUUGACAUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUCGAAAUAUUGAUUAGGAAAGGCAACAGCAAAAUAAAUACAA ------------------...............((((...((((.(((........))).))))))))....(((((((((....)))))..)))).(....)................ ( -17.10) >DroEre_CAF1 42244 115 + 1 AACUA----UAUAAAGGAAUAAAACAAAUUAAACUGACCUUGACUUCCCCAGAUCAGGAAGUCAUCAGAAAGUUCCAUCAAAAUAUUGAUUAGGAAAGGCUACAGCUAAAUAAGCACAA .....----........................((((...((((((((........))))))))))))....(((((((((....)))))..))))........(((.....))).... ( -22.10) >DroYak_CAF1 43389 119 + 1 AACUACAUAUAUUAAGGAAUAAAACAUAUUGAACUGACAUUGACUUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUCGAAAUAUUGAUUAGGAAAGGCUACAGCAAAAUAAAUACAA ..(((..........(((((....(.....)..((((...((((((((........))))))))))))...)))))(((((....)))))))).....((....))............. ( -21.70) >consensus A_CUA____U_U__AG_AAUAAAACAAAUUAAACUGACCUUGACUUCCAGAGAUCAGGAAGUCAUCAGAAAGUUCCAUCGAAAUAUUGAUUAGGAAAGGCUACAGCAAAAUAAAUACAA .................................((((...((((((((........))))))))))))....(((((((((....)))))..))))..((....))............. (-19.54 = -19.78 + 0.24)



| Location | 4,805,935 – 4,806,050 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -17.52 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4805935 115 - 27905053 UUGUAUUUAUUUUGCUGUAGCCUUUCCUAAUCAAUAUUUCGAUGGAACUUUCUGAUGACUUCCUGAUCUCUGGAAGUCAAGAUCAGUUUAAUUUGUUUUAUUUCUGUACA----UAGCU .............(((((.....((((..(((........)))))))....((((((((((((........))))))))...)))).......................)----)))). ( -22.80) >DroSec_CAF1 41246 101 - 1 UUGUAUUUAUUUUGCUGUUGCCUUUCCUAAUCAAUAUUUCGAUGGAACUUUCUGAUGACUUCCUGGUCUCUGGAUGUCAAGAUCAGUUUAAUAUGUUUUAU------------------ .......((((..((((......((((..(((........)))))))...(((..((((.(((........))).))))))).))))..))))........------------------ ( -16.90) >DroSim_CAF1 37460 101 - 1 UUGUAUUUAUUUUGCUGUUGCCUUUCCUAAUCAAUAUUUCGAUGGAACUUUCUGAUGACUUCCUGAUCUCUGGAUGUCAAGCUCAGUUUAAUUUGUUUUAU------------------ ....((..(((..((((..((..((((..(((........)))))))........((((.(((........))).)))).)).))))..)))..)).....------------------ ( -17.50) >DroEre_CAF1 42244 115 - 1 UUGUGCUUAUUUAGCUGUAGCCUUUCCUAAUCAAUAUUUUGAUGGAACUUUCUGAUGACUUCCUGAUCUGGGGAAGUCAAGGUCAGUUUAAUUUGUUUUAUUCCUUUAUA----UAGUU ....((..(((.(((((..((((((((..(((((....)))))))))........(((((((((......)))))))))))))))))).)))..))..............----..... ( -27.90) >DroYak_CAF1 43389 119 - 1 UUGUAUUUAUUUUGCUGUAGCCUUUCCUAAUCAAUAUUUCGAUGGAACUUUCUGAUGACUUCCUGAUCUCUGGAAGUCAAUGUCAGUUCAAUAUGUUUUAUUCCUUAAUAUAUGUAGUU .............((....)).....(((....(((((..((((((((...((((((((((((........))))))))...))))........))))))))....)))))...))).. ( -21.10) >consensus UUGUAUUUAUUUUGCUGUAGCCUUUCCUAAUCAAUAUUUCGAUGGAACUUUCUGAUGACUUCCUGAUCUCUGGAAGUCAAGAUCAGUUUAAUUUGUUUUAUU_CU__A_A____UAG_U ....((..(((..((((......((((..(((........)))))))........((((((((........))))))))....))))..)))..))....................... (-17.52 = -17.60 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:38 2006