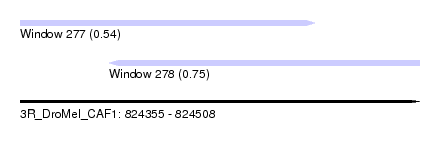
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 824,355 – 824,508 |
| Length | 153 |
| Max. P | 0.750342 |

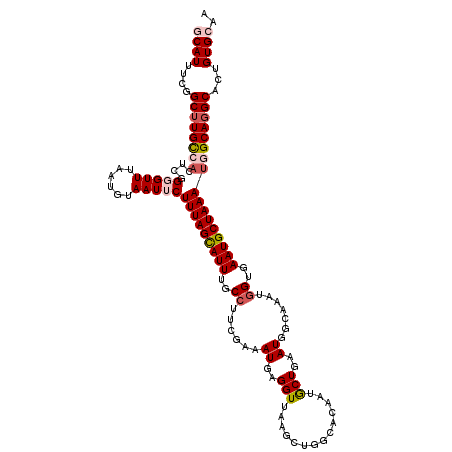
| Location | 824,355 – 824,468 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -21.74 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 824355 113 + 27905053 CCCAGAA------AUAACUGCUUGAGGUGACCCAUUUCGGUUGCACAGUGCCUGCCAUUUAGCAUUCACCAUUUGCCAUUCAGCAUUGUGCCAGCUUAACCUCAUUUCGAA-GCAAAUGC .......------.....((((((((((((........(((((((((((((.((.......(((.........)))....)))))))))).))))).....))))))).))-)))..... ( -28.22) >DroSec_CAF1 6648 113 + 1 CCCUGAA------AUAUUUGCUUGAGGUGACCCAUUUCGGUUGCACAGUGCCUGCCAUUUAGCAUUCACCAUUUG-CAUUCAGCAUUGUGCCAGCUUAACCUCAUUUCGAAGGCAAAUGC .......------.((((((((((((((((((......))))(((((((((.((.......(((.........))-)...))))))))))).......)))))).......)))))))). ( -31.11) >DroSim_CAF1 3828 114 + 1 CCCUAAA------AUAUCUGCUUGAGGUGACCCAUUUCGGUUGCACAGUGCCUGCCAUUUAGCAUUCACCAUUUGCCAUUCAGCAUUGUGCCAGCUUAACCUCAUUUCGAAGGCAAAUGC .......------.(((.((((((((((((((......))))(((((((((.((.......(((.........)))....))))))))))).......)))))).......)))).))). ( -28.01) >DroEre_CAF1 7108 114 + 1 CACUGAU------AUAUCUGCUUGAGCUGACCCAUUUCGGUUGCACAGUGCCUGCUAUUUAGCAUUCACCAUUGGCUAUUCAGUAUUGUGUCAGCUUAACCCCAUUUCGAAGGCAAAUAC .......------.(((.(((((((((((((.((.((..((.((.(((((..((((....)))).....))))))).))..))...)).))))))))).............)))).))). ( -25.01) >DroYak_CAF1 6744 120 + 1 CACUGAUACUCGUAUAUCUGUUUGAGGUGACCCAUUUUGCUUGCACAGUGCCUGCUAUUUAGCAUUCACCAUUGGCCAUUCAGCAUUGUGCCAGCGUAACCUUAUUUCGAAGGCAAAUAC ...........((((.(.(.((((((((((.......((((.(((((((((.((((....))))........(((....)))))))))))).)))).....)))))))))).).).)))) ( -29.20) >consensus CCCUGAA______AUAUCUGCUUGAGGUGACCCAUUUCGGUUGCACAGUGCCUGCCAUUUAGCAUUCACCAUUUGCCAUUCAGCAUUGUGCCAGCUUAACCUCAUUUCGAAGGCAAAUGC ..............(((.((((((((((((........(((((((((((((.(((......)))....(.....).......)))))))).))))).....)))))))))..))).))). (-21.74 = -22.02 + 0.28)

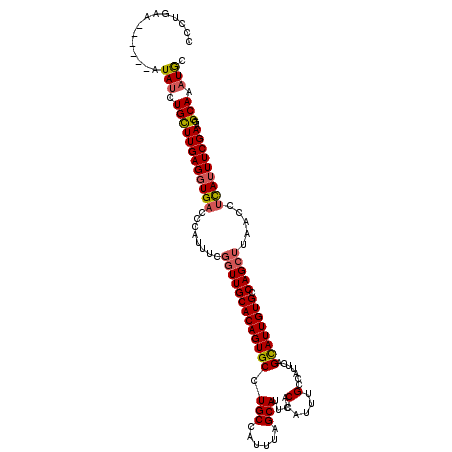
| Location | 824,389 – 824,508 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -27.58 |
| Energy contribution | -28.82 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 824389 119 - 27905053 GCAUUUCGGCUUGUCACUCGGGGUUUAAUGUAAUUCUUUAGCAUUUGC-UUCGAAAUGAGGUUAAGCUGGCACAAUGCUGAAUGGCAAAUGGUGAAUGCUAAAUGGCAGGCACUGUGCAA ......((((((((((.(((((((..(((((.........))))).))-)))))..)))...)))))))((((..((((....))))...((((..((((....))))..)))))))).. ( -37.80) >DroSec_CAF1 6682 119 - 1 GCAUUUCGGCUUGCCACUCGGGGUUUAAUGUAAUUCUUUAGCAUUUGCCUUCGAAAUGAGGUUAAGCUGGCACAAUGCUGAAUG-CAAAUGGUGAAUGCUAAAUGGCAGGCACUGUGCAA ((((....((((((((...((((((......))))))((((((((..(((((.....))).....((.(((.....)))....)-)....))..)))))))).))))))))...)))).. ( -40.10) >DroSim_CAF1 3862 120 - 1 GCAUUUCGGCUUGCCACUCGGGGUUUAAUGUAAUUCUUUAGCAUUUGCCUUCGAAAUGAGGUUAAGCUGGCACAAUGCUGAAUGGCAAAUGGUGAAUGCUAAAUGGCAGGCACUGUGCAA ((((....((((((((...((((((......))))))((((((((..(((((.....))).....((((((.....)))....)))....))..)))))))).))))))))...)))).. ( -40.90) >DroEre_CAF1 7142 120 - 1 CCAUUUUGGCUUGCCACUCGGUGUUUAAUGUAAUUCUUUAGUAUUUGCCUUCGAAAUGGGGUUAAGCUGACACAAUACUGAAUAGCCAAUGGUGAAUGCUAAAUAGCAGGCACUGUGCAA ......(((((......((((((((...(((.......((((....(((((......)))))...))))))).))))))))..)))))((((((..((((....))))..)))))).... ( -28.61) >DroYak_CAF1 6784 120 - 1 CCAUUUCGGCAUGCCUCUGUGUGUUUAAUGUAAUUCUUUAGUAUUUGCCUUCGAAAUAAGGUUACGCUGGCACAAUGCUGAAUGGCCAAUGGUGAAUGCUAAAUAGCAGGCACUGUGCAA (((((.(((((((((...(((((.......(((....)))......(((((......)))))))))).))))...))))))))))...((((((..((((....))))..)))))).... ( -33.90) >consensus GCAUUUCGGCUUGCCACUCGGGGUUUAAUGUAAUUCUUUAGCAUUUGCCUUCGAAAUGAGGUUAAGCUGGCACAAUGCUGAAUGGCAAAUGGUGAAUGCUAAAUGGCAGGCACUGUGCAA ((((....((((((((....(((((......)))))(((((((((..((......((..(((..............)))..)).......))..)))))))))))))))))...)))).. (-27.58 = -28.82 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:36 2006