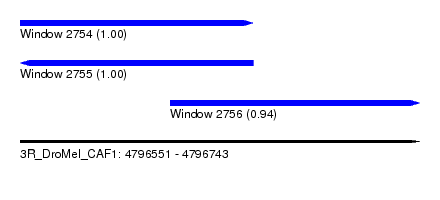
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,796,551 – 4,796,743 |
| Length | 192 |
| Max. P | 0.999986 |

| Location | 4,796,551 – 4,796,663 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4796551 112 + 27905053 ---UGUUGUUGUUGUGGCUCGGCAUGGUAAUGAGCUGAGCAGCCAAGAAACAGAAACAGAGUCUGAUUAUCAUGAGUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAA ---...........((((((..((((((((((.(((....))))......((((.......))))))))))))).((((((((....)))))))))))))).............. ( -31.30) >DroSec_CAF1 31915 109 + 1 ---UGUU---GUUGUGGCUCGGCAUGGUAAUGAGCUGAGCAGCCAAGAAACAGAAACAGAGUCUGAUUAUCAUGAGUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAA ---....---....((((((..((((((((((.(((....))))......((((.......))))))))))))).((((((((....)))))))))))))).............. ( -31.30) >DroSim_CAF1 27970 112 + 1 ---UGUUGUUGUUGUGGCUCGGCAUGGUAAUGAGCUGAGCAGCCAAGAAACAGAAACAGAGUCUGAUUAUCAUGAGUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAA ---...........((((((..((((((((((.(((....))))......((((.......))))))))))))).((((((((....)))))))))))))).............. ( -31.30) >DroEre_CAF1 32672 115 + 1 AGUUGUUUUUGUUGUGGCUCGGCAUGGUAAUGAGCUGAGCAGCCAAGAAACAGAAACAGAGUCUGAUUAUCAUGAGUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAA ..(..((((((((.((((((..((((((((((.(((....))))......((((.......))))))))))))).((((((((....)))))))))))))).)).))))))..). ( -35.30) >DroYak_CAF1 33096 115 + 1 AGUUGUUUUUGCUGUGGCUCGGCAUGGUAAUGAGCUGAGCAGCCAAGAAACAGAAACAGAGUCUGAUUAUCAUGAGUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAA ..(..((((((((.((((((..((((((((((.(((....))))......((((.......))))))))))))).((((((((....))))))))))))))..))))))))..). ( -40.40) >consensus ___UGUUGUUGUUGUGGCUCGGCAUGGUAAUGAGCUGAGCAGCCAAGAAACAGAAACAGAGUCUGAUUAUCAUGAGUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAA ..............((((((..((((((((((.(((....))))......((((.......))))))))))))).((((((((....)))))))))))))).............. (-31.30 = -31.30 + 0.00)



| Location | 4,796,551 – 4,796,663 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.41 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4796551 112 - 27905053 UUCACUUUUACUCUCGGCUCAUAAGCAUAAUCAUGCUUACUCAUGAUAAUCAGACUCUGUUUCUGUUUCUUGGCUGCUCAGCUCAUUACCAUGCCGAGCCACAACAACAACA--- ...............(((((.(((((((....)))))))..((((.((((((((.......))))......((((....)))).)))).))))..)))))............--- ( -24.40) >DroSec_CAF1 31915 109 - 1 UUCACUUUUACUCUCGGCUCAUAAGCAUAAUCAUGCUUACUCAUGAUAAUCAGACUCUGUUUCUGUUUCUUGGCUGCUCAGCUCAUUACCAUGCCGAGCCACAAC---AACA--- ...............(((((.(((((((....)))))))..((((.((((((((.......))))......((((....)))).)))).))))..))))).....---....--- ( -24.40) >DroSim_CAF1 27970 112 - 1 UUCACUUUUACUCUCGGCUCAUAAGCAUAAUCAUGCUUACUCAUGAUAAUCAGACUCUGUUUCUGUUUCUUGGCUGCUCAGCUCAUUACCAUGCCGAGCCACAACAACAACA--- ...............(((((.(((((((....)))))))..((((.((((((((.......))))......((((....)))).)))).))))..)))))............--- ( -24.40) >DroEre_CAF1 32672 115 - 1 UUCACUUUUACUCUCGGCUCAUAAGCAUAAUCAUGCUUACUCAUGAUAAUCAGACUCUGUUUCUGUUUCUUGGCUGCUCAGCUCAUUACCAUGCCGAGCCACAACAAAAACAACU ...............(((((.(((((((....)))))))..((((.((((((((.......))))......((((....)))).)))).))))..)))))............... ( -24.40) >DroYak_CAF1 33096 115 - 1 UUCACUUUUACUCUCGGCUCAUAAGCAUAAUCAUGCUUACUCAUGAUAAUCAGACUCUGUUUCUGUUUCUUGGCUGCUCAGCUCAUUACCAUGCCGAGCCACAGCAAAAACAACU ...............(((((.(((((((....)))))))..((((.((((((((.......))))......((((....)))).)))).))))..)))))............... ( -24.40) >consensus UUCACUUUUACUCUCGGCUCAUAAGCAUAAUCAUGCUUACUCAUGAUAAUCAGACUCUGUUUCUGUUUCUUGGCUGCUCAGCUCAUUACCAUGCCGAGCCACAACAAAAACA___ ...............(((((.(((((((....)))))))..((((.((((((((.......))))......((((....)))).)))).))))..)))))............... (-24.40 = -24.40 + 0.00)



| Location | 4,796,623 – 4,796,743 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -29.32 |
| Energy contribution | -29.27 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4796623 120 + 27905053 GUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAAAGAGGCGGAGAAUGUGUGUGUAAAGAAUGCCUUGAGUGUCUGAGUGCCUCGUAUGUAGGCCAAGUCUAUUAUGCGGCUAG ((((((((....)))))))).(((((((.(((..((.....((((((....................))))))......))...))))))((((((((((...))))).))))))))).. ( -34.05) >DroSec_CAF1 31984 116 + 1 GUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAAAGAGGCGGAGAGUG----UGUAAAGAAUGCCUUGAGUGUCUGAGUGCCUCGUAUGUAGGCCAAGUCUAUUAUGCGGCUAG ((((((((....)))))))).(((((((.(((..((.....((((((.......----.........))))))......))...))))))((((((((((...))))).))))))))).. ( -34.29) >DroSim_CAF1 28042 118 + 1 GUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAAAGAGGCGGAGAGUG--UGUGUAAAGAAUGCCUUGAGUGUCUGAGUGCCUCGUAUGUAGGCCAAGUCUAUUAUGCGGCUAG ((((((((....)))))))).(((((((.(((........(((.((...(((.(--(((.......)))))))..)).)))...))))))((((((((((...))))).))))))))).. ( -34.60) >DroEre_CAF1 32747 118 + 1 GUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAAAGAGGCGGAGAGUG--UGUGUAAAGAAUGCCUUGAGUGUCUGAGUGCCUCGUAUGUAGGCCAAGUCUAUUAUGCGGCUAG ((((((((....)))))))).(((((((.(((........(((.((...(((.(--(((.......)))))))..)).)))...))))))((((((((((...))))).))))))))).. ( -34.60) >DroYak_CAF1 33171 118 + 1 GUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAAAGAGGCGGAGAGUG--UGUGUAAAGAAUGCCUUGAGUGUCUGAGUGCCUCGUAUGUAGGCCAAGUCUAUUAUGCGGCUAG ((((((((....)))))))).(((((((.(((........(((.((...(((.(--(((.......)))))))..)).)))...))))))((((((((((...))))).))))))))).. ( -34.60) >DroAna_CAF1 31654 116 + 1 GUAAGCAUGAUUAUGCUUAUGAGCAGAGAGUAAAAGUGACUGACGGAACGAGCG--AGUG-AAAGAAUGCCUUGAGUGUCUGAGUGUUUUGUAUGUAGGCCAAGUCUAUUAUGCGGCUA- ((((((((....)))))))).(((.............(((...((((.((..((--((.(-........)))))..))))))...))).(((((((((((...))))).))))))))).- ( -25.30) >consensus GUAAGCAUGAUUAUGCUUAUGAGCCGAGAGUAAAAGUGAAAGAGGCGGAGAGUG__UGUGUAAAGAAUGCCUUGAGUGUCUGAGUGCCUCGUAUGUAGGCCAAGUCUAUUAUGCGGCUAG ((((((((....)))))))).(((((((.(((..((.....((((((....................))))))......))...))))))((((((((((...))))).))))))))).. (-29.32 = -29.27 + -0.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:27 2006